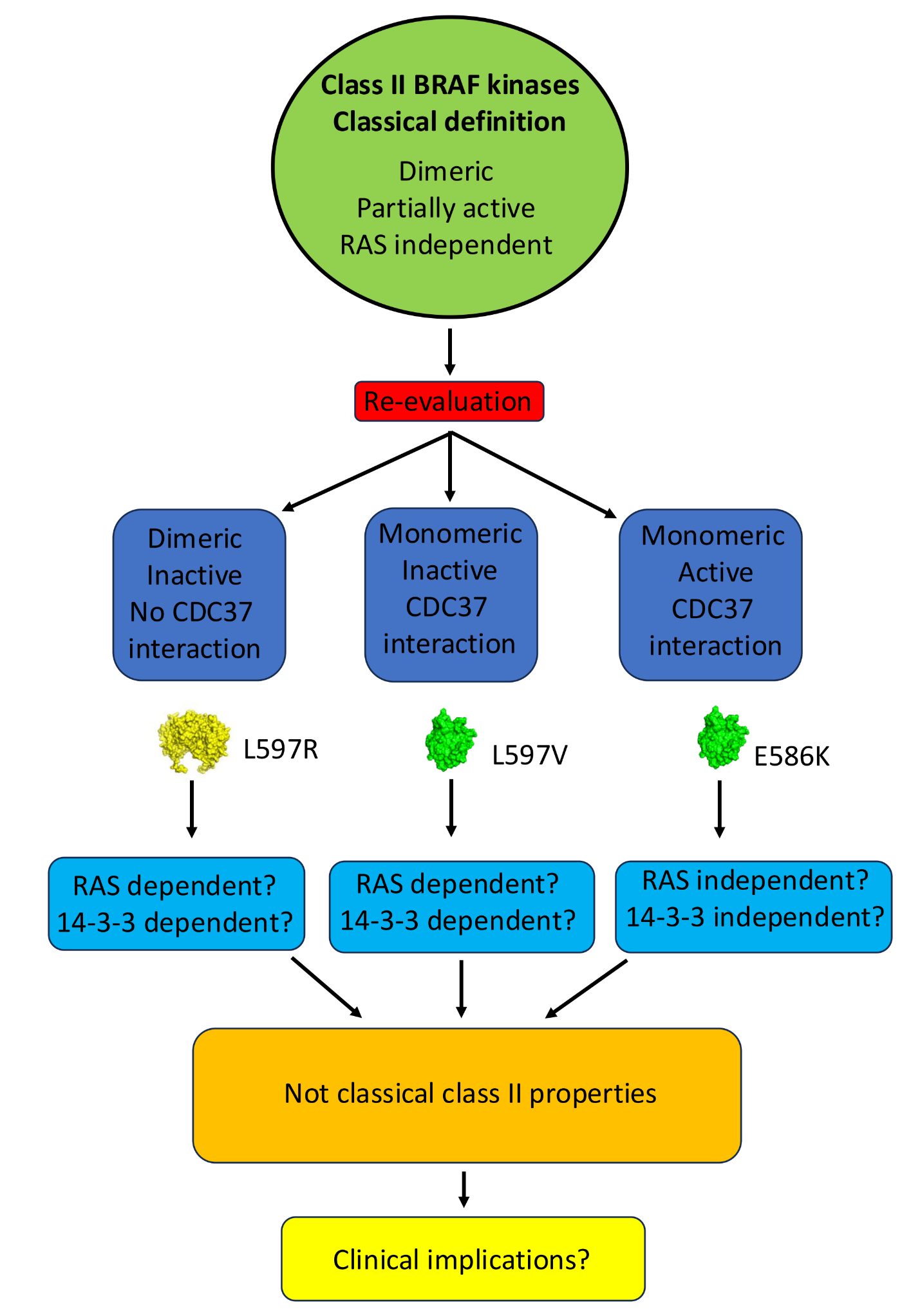
Oncogenic BRAF mutations represent a variety of amino acid changes at multiple locations within the BRAF kinase domain. Their classification, as class I, II or III, depends on their RAS dependency, their dimerization status in their signalling state and kinase activity. However, many of these mutants have not been fully characterised for their in vitro oligomeric state and kinase activity. Additionally, their interaction with the Hsp90-CDC37 complex, an important component in their stability and activation, is poorly defined. As these properties likely affect the exact mechanism by which these mutants drive carcinogenesis it is of clinical importance that such a characterization is undertaken. Here we report the purification and characterization of a select number of BRAF-mutant proteins so that we can better understand their mode of action. We find that the purified class-II E586K-mutant kinase domain of BRAF is active in its monomeric state, whereas the class-II mutant L597V is, in contrast, an inactive monomer. We also find that purified T599W, where the tryptophan substitution eliminates an activating-phosphorylation site, was active for kinase activity. Consistent with the monomeric state of these mutant BRAF proteins, they interacted with CDC37, although T599W did show somewhat reduced affinity. Our findings, question the strict classification of specific BRAF mutants and suggest further investigations are required to fully understand their carcinogenic mechanism.
- Open Access
- Article
Analyses of a Select Set of BRAF Mutants and Implications for Their Mechanistic Action as Drivers of Carcinogenesis
Author Information
Received: 09 Aug 2025 | Revised: 27 Sep 2025 | Accepted: 29 Sep 2025 | Published: 11 Oct 2025
Abstract
Graphical Abstract
References
- 1.Lemmon, M.A.; Schlessinger, J. Cell signaling by receptor tyrosine kinases. Cell 2010, 141, 1117–1134. https://doi.org/10.1016/j.cell.2010.06.011.
- 2.Huse, M.; Kuriyan, J. The conformational plasticity of protein kinases. Cell 2002, 109, 275–282. https://doi.org/10.1016/s0092-8674(02)00741-9.
- 3.Bharti, J.; Gogu, P.; Pandey, S.K.; et al. BRAF V600E in cancer: Exploring structural complexities, mutation profiles, and pathway dysregulation. Exp. Cell Res. 2025, 446, 114440. https://doi.org/10.1016/j.yexcr.2025.114440.
- 4.Keramisanou, D.; Aboalroub, A.; Zhang, Z.; et al. Molecular Mechanism of Protein Kinase Recognition and Sorting by the Hsp90 Kinome-Specific Cochaperone Cdc37. Mol. Cell 2016, 62, 260–271. https://doi.org/10.1016/j.molcel.2016.04.005.
- 5.Xu, H. ATP-Driven Nonequilibrium Activation of Kinase Clients by the Molecular Chaperone Hsp90. Biophys. J. 2020, 119, 1538–1549. https://doi.org/10.1016/j.bpj.2020.08.038.
- 6.Hunter, T.; Poon, R.Y.C. Cdc37: A protein kinase chaperone? Trends Cell Biol. 1997, 7, 157–161.
- 7.Wartmann, M.; Davis, R.J. The native structure of the activated Raf protein kinase is a membrane-bound multi-subunit complex. J. Biol. Chem. 1994, 269, 6695–6701.
- 8.Nussinov, R.; Tsai, C.J.; Jang, H. Does Ras Activate Raf and PI3K Allosterically? Front. Oncol. 2019, 9, 1231. https://doi.org/10.3389/fonc.2019.01231.
- 9.Grbovic, O.M.; Basso, A.D.; Sawai, A.; et al. V600E B-Raf requires the Hsp90 chaperone for stability and is degraded in response to Hsp90 inhibitors. Proc. Natl. Acad. Sci. USA 2006, 103, 57–62. https://doi.org/10.1073/pnas.0609973103.
- 10.Sharp, S.; Workman, P. Inhibitors of the HSP90 molecular chaperone: Current status. Adv. Cancer Res. 2006, 95, 323–348. https://doi.org/10.1016/S0065-230X(06)95009-X.
- 11.Schulte, T.W.; Blagosklonny, M.V.; Ingui, C.; et al. Disruption of the Raf-1-Hsp90 molecular complex results in destabilization of Raf-1 and loss of Raf-1-Ras association. J. Biol. Chem. 1995, 270, 24585–24588.
- 12.Polier, S.; Samant, R.S.; Clarke, P.A.; et al. ATP-competitive inhibitors block protein kinase recruitment to the Hsp90-Cdc37 system. Nat. Chem. Biol. 2013, 9, 307–312. https://doi.org/10.1038/nchembio.1212.
- 13.Kohler, M.; Brummer, T. B-Raf activation loop phosphorylation revisited. Cell Cycle 2016, 15, 1171–1173. https://doi.org/10.1080/15384101.2016.1159111.
- 14.Lavoie, H.; Therrien, M. Regulation of RAF protein kinases in ERK signalling. Nat. Reviews. Mol. Cell Biol. 2015, 16, 281–298. https://doi.org/10.1038/nrm3979.
- 15.Zhang, B.H.; Guan, K.L. Activation of B-Raf kinase requires phosphorylation of the conserved residues Thr598 and Ser601. EMBO J. 2000, 19, 5429–5439. https://doi.org/10.1093/emboj/19.20.5429.
- 16.Kornev, A.P.; Taylor, S.S.; Ten Eyck, L.F. A helix scaffold for the assembly of active protein kinases. Proc. Natl. Acad. Sci. USA 2008, 105, 14377–14382. https://doi.org/10.1073/pnas.0807988105.
- 17.Hu, J.; Ahuja, L.G.; Meharena, H.S.; et al. Kinase regulation by hydrophobic spine assembly in cancer. Mol. Cell Biol. 2015, 35, 264–276. https://doi.org/10.1128/MCB.00943-14.
- 18.Kim, J.; Ahuja, L.G.; Chao, F.A.; et al. A dynamic hydrophobic core orchestrates allostery in protein kinases. Sci. Adv. 2017, 3, e1600663. https://doi.org/10.1126/sciadv.1600663.
- 19.Taylor, S.S.; Kornev, A.P. Protein kinases: Evolution of dynamic regulatory proteins. Trends Biochem. Sci. 2011, 36, 65–77. https://doi.org/10.1016/j.tibs.2010.09.006.
- 20.Diedrich, B.; Rigbolt, K.T.; Roring, M.; et al. Discrete cytosolic macromolecular BRAF complexes exhibit distinct activities and composition. EMBO J. 2017, 36, 646–663. https://doi.org/10.15252/embj.201694732.
- 21.Cook, F.A.; Cook, S.J. Inhibition of RAF dimers: It takes two to tango. Biochem. Soc. Trans. 2021, 49, 237–251. https://doi.org/10.1042/BST20200485.
- 22.Yao, Z.; Torres, N.M.; Tao, A.; et al. BRAF Mutants Evade ERK-Dependent Feedback by Different Mechanisms that Determine Their Sensitivity to Pharmacologic Inhibition. Cancer Cell 2015, 28, 370–383. https://doi.org/10.1016/j.ccell.2015.08.001.
- 23.Yao, Z.; Yaeger, R.; Rodrik-Outmezguine, V.S.; et al. Tumours with class 3 BRAF mutants are sensitive to the inhibition of activated RAS. Nature 2017, 548, 234–238. https://doi.org/10.1038/nature23291.
- 24.Karoulia, Z.; Gavathiotis, E.; Poulikakos, P.I. New perspectives for targeting RAF kinase in human cancer. Nature reviews. Cancer 2017, 17, 676–691. https://doi.org/10.1038/nrc.2017.79.
- 25.Kordes, M.; Roring, M.; Heining, C.; et al. Cooperation of BRAF(F595L) and mutant HRAS in histiocytic sarcoma provides new insights into oncogenic BRAF signaling. Leukemia 2016, 30, 937–946. https://doi.org/10.1038/leu.2015.319.
- 26.Bjorklund, D.M.; Morgan, R.M.L.; Oberoi, J.; et al. Recognition of BRAF by CDC37 and Re-Evaluation of the Activation Mechanism for the Class 2 BRAF-L597R Mutant. Biomolecules 2022, 12, 905. https://doi.org/10.3390/biom12070905.
- 27.Perdew, G.H.; Wiegand, H.; VandenHeuvel, J.P.; et al. A 50 kilodalton protein associated with raf and pp(60v-src) protein kinases is a mammalian homolog of the cell cycle control protein cdc37. Biochemistry 1997, 36, 3600–3607.
- 28.Wan, X.; Yap, J.; Chen, J.; et al. Oncogenic non-V600 mutations evade the regulatory machinery of RAF including the Cdc37/Hsp90 chaperone and the 14-3-3 scaffold. Theranostics 2025, 15, 2035–2051. https://doi.org/10.7150/thno.103958.
- 29.Lauinger, M.; Christen, D.; Klar, R.F.U.; et al. BRAF(Deltabeta3-alphaC) in-frame deletion mutants differ in their dimerization propensity, HSP90 dependence, and druggability. Sci. Adv. 2023, 9, eade7486. https://doi.org/10.1126/sciadv.ade7486.
- 30.Zhao, Y.; Yu, H.; Ida, C.M.; et al. Assessment of RAS Dependency for BRAF Alterations Using Cancer Genomic Databases. JAMA Netw. Open 2021, 4, e2035479. https://doi.org/10.1001/jamanetworkopen.2020.35479.
- 31.Anastasaki, C.; Orozco, P.; Gutmann, D.H. RAS and beyond: The many faces of the neurofibromatosis type 1 protein. Dis. Model. Mech. 2022, 15, dmm049362. https://doi.org/10.1242/dmm.049362.
- 32.Liau, N.P.D.; Venkatanarayan, A.; Quinn, J.G.; et al. Dimerization Induced by C-Terminal 14-3-3 Binding Is Sufficient for BRAF Kinase Activation. Biochemistry 2020, 59, 3982–3992. https://doi.org/10.1021/acs.biochem.0c00517.
- 33.Liau, N.P.D.; Wendorff, T.J.; Quinn, J.G.; et al. Negative regulation of RAF kinase activity by ATP is overcome by 14-3-3-induced dimerization. Nat. Struct. Mol. Biol. 2020, 27, 134–141. https://doi.org/10.1038/s41594-019-0365-0.
- 34.Kondo, Y.; Ognjenovic, J.; Banerjee, S.; et al. Cryo-EM structure of a dimeric B-Raf:14-3-3 complex reveals asymmetry in the active sites of B-Raf kinases. Science 2019, 366, 109–115. https://doi.org/10.1126/science.aay0543.
- 35.Heidorn, S.J.; Milagre, C.; Whittaker, S.; et al. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell 2010, 140, 209–221. https://doi.org/10.1016/j.cell.2009.12.040.
- 36.Poulikakos, P.I.; Zhang, C.; Bollag, G.; et al. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature 2010, 464, 427–430. https://doi.org/10.1038/nature08902.
- 37.Hatzivassiliou, G.; Song, K.; Yen, I.; et al. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature 2010, 464, 431–435. https://doi.org/10.1038/nature08833.
- 38.Garnett, M.J.; Rana, S.; Paterson, H.; et al. Wild-type and mutant B-RAF activate C-RAF through distinct mechanisms involving heterodimerization. Mol. Cell 2005, 20, 963–969. https://doi.org/10.1016/j.molcel.2005.10.022.
- 39.Johnson, D.B.; Dahlman, K.B. Class Matters: Sensitivity of BRAF-Mutant Melanoma to MAPK Inhibition. Clin. Cancer Res. 2018, 24, 6107–6109. https://doi.org/10.1158/1078-0432.CCR-18-1795.
- 40.Baik, C.S.; Myall, N.J.; Wakelee, H.A. Targeting BRAF-Mutant Non-Small Cell Lung Cancer: From Molecular Profiling to Rationally Designed Therapy. Oncologist 2017, 22, 786–796. https://doi.org/10.1634/theoncologist.2016-0458.
- 41.Mikhailenko, D.S.; Efremov, G.D.; Safronova, N.Y.; et al. Detection of Rare Mutations by Routine Analysis of KRAS, NRAS, and BRAF Oncogenes. Bull. Exp. Biol. Med. 2017, 162, 375–378. https://doi.org/10.1007/s10517-017-3619-z.
- 42.Center, V.-I.C. My Cancer Genome. Available online: https://www.mycancergenome.org/content/alteration/braf-l597r (accessed on 8 July 2025).
- 43.Dankner, M.; Rose, A.A.N.; Rajkumar, S.; et al. Classifying BRAF alterations in cancer: New rational therapeutic strategies for actionable mutations. Oncogene 2018, 37, 3183–3199. https://doi.org/10.1038/s41388-018-0171-x.
- 44.Shackelford, R.; Pollen, M.; Vora, M.; et al. Malignant Melanoma with Concurrent BRAF E586K and NRAS Q81K Mutations. Case Rep. Oncol. 2014, 7, 297–300. https://doi.org/10.1159/000362788.
- 45.Swofford, B.P.; Homsi, J. Uncommon BRAF Mutations Associated with Durable Response to Immunotherapy in Patients with Metastatic Melanoma. Case Rep. Oncol. Med. 2017, 2017, 8241624. https://doi.org/10.1155/2017/8241624.
- 46.Lei, L.; Wang, W.X.; Zhu, Y.C.; et al. Association between BRAF mutant classification and the efficacy of pemetrexed-based chemotherapy in Chinese advanced non-small cell lung cancer patients: A multicenter retrospective study. Transl. Cancer Res. 2020, 9, 6039–6049. https://doi.org/10.21037/tcr-20-480.
- 47.Center, V.-I.C. My Cancer Genome. Available online: https://www.mycancergenome.org/content/alteration/braf-e586k/ (accessed on 8 July 2025).
- 48.Noeparast, A.; Teugels, E.; Giron, P.; et al. Non-V600 BRAF mutations recurrently found in lung cancer predict sensitivity to the combination of Trametinib and Dabrafenib. Oncotarget 2017, 8, 60094–60108. https://doi.org/10.18632/oncotarget.11635.
- 49.Liu, J.; Xie, H. BRAF Non-V600 Mutations in Metastatic Colorectal Cancer. Cancers 2023, 15, 4604. https://doi.org/10.3390/cancers15184604.
- 50.Cremolini, C.; Di Bartolomeo, M.; Amatu, A.; et al. BRAF codons 594 and 596 mutations identify a new molecular subtype of metastatic colorectal cancer at favorable prognosis. Ann. Oncol. 2015, 26, 2092–2097. https://doi.org/10.1093/annonc/mdv290.
- 51.Chao, W.R.; Lee, Y.J.; Lee, M.Y.; et al. High frequency of BRAF mutations in primary mucinous ovarian carcinoma of Taiwanese patients. Taiwan. J. Obstet. Gynecol. 2021, 60, 1072–1077. https://doi.org/10.1016/j.tjog.2021.09.019.
- 52.Tsai, J.; Lee, J.T.; Wang, W.; et al. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proc. Natl. Acad. Sci. USA 2008, 105, 3041–3046. https://doi.org/10.1073/pnas.0711741105.
- 53.Andreadi, C.; Cheung, L.K.; Giblett, S.; et al. The intermediate-activity (L597V)BRAF mutant acts as an epistatic modifier of oncogenic RAS by enhancing signaling through the RAF/MEK/ERK pathway. Genes. Dev. 2012, 26, 1945–1958. https://doi.org/10.1101/gad.193458.112.
- 54.Dahlman, K.B.; Xia, J.; Hutchinson, K.; et al. BRAF(L597) mutations in melanoma are associated with sensitivity to MEK inhibitors. Cancer Discov. 2012, 2, 791–797. https://doi.org/10.1158/2159-8290.CD-12-0097.
- 55.Davies, H.; Bignell, G.R.; Cox, C.; et al. Mutations of the BRAF gene in human cancer. Nature 2002, 417, 949–954. https://doi.org/10.1038/nature00766.
- 56.Wan, P.T.; Garnett, M.J.; Roe, S.M.; et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell 2004, 116, 855–867. https://doi.org/10.1016/s0092-8674(04)00215-6.
- 57.Woolley, C.E.; Domingo, E.; Fernandez-Tajes, J.; et al. Coevolution of Atypical BRAF and KRAS Mutations in Colorectal Tumorigenesis. Mol. Cancer Res. 2025, 23, 300–312. https://doi.org/10.1158/1541-7786.MCR-24-0464.
- 58.Dankner, M.; Lajoie, M.; Moldoveanu, D.; et al. Dual MAPK Inhibition Is an Effective Therapeutic Strategy for a Subset of Class II BRAF Mutant Melanomas. Clin. Cancer Res. 2018, 24, 6483–6494. https://doi.org/10.1158/1078-0432.CCR-17-3384.
Issue
Volume 1, Issue 1How to Cite
Bjorklund, D. M.; Jeanne, X.; Prodromou, C. Analyses of a Select Set of BRAF Mutants and Implications for Their Mechanistic Action as Drivers of Carcinogenesis. Biomolecular Mechanisms and Innovations 2026, 1 (1), 2.
RIS
BibTex
Copyright & License

Copyright (c) 2025 by the authors.
This work is licensed under a Creative Commons Attribution 4.0 International License.
Contents
References


