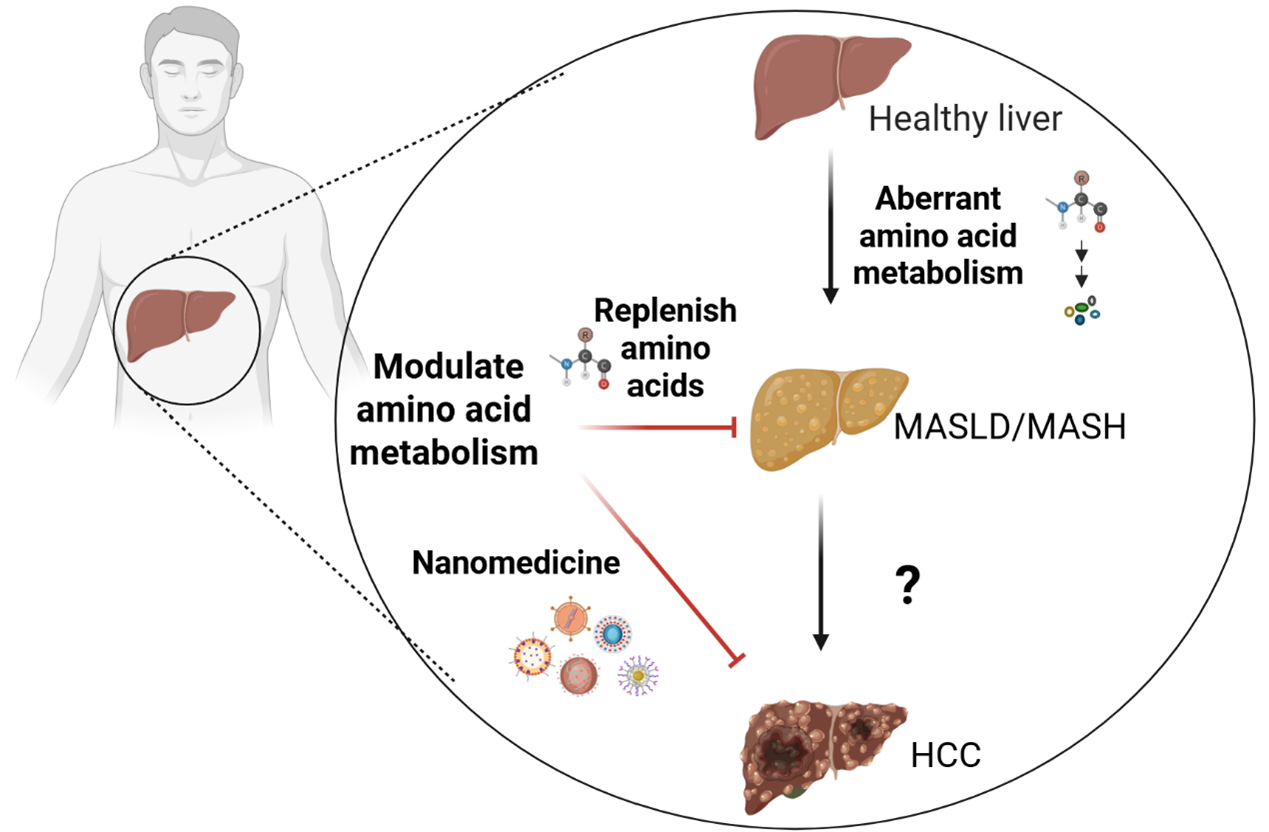
Metabolic dysfunction-associated steatotic liver disease (MASLD) has been proposed as a more precise term to characterize steatosis amid metabolic dysregulation and is projected to emerge as the predominant cause of hepatocellular carcinoma (HCC) globally, given the rising prevalence of metabolic comorbidities. Particularly in its inflammatory form, known as metabolic dysfunction-associated steatohepatitis (MASH), hepatic metabolism is profoundly altered. Amino acids are fundamental building blocks that support cellular metabolism and biosynthesis, alongside monosaccharides and fatty acids. Emerging research suggests that aberrant amino acid metabolism in MASLD/MASH and HCC impacts mitochondrial function and redox equilibrium. Nonetheless, the involvement of amino acid metabolism in the progression from MASLD/MASH to HCC is still inadequately comprehended. This review summarizes the aberrant amino acid metabolism in MASLD/MASH and HCC, as well as nanomedicine-based approaches for modulating this metabolism to facilitate the discovery of more effective biomarkers and precision therapeutics for the prevention of MASLD/MASH and HCC.
- Open Access
- Review
Nanomedicine-Based Strategies for Regulating Abnormal Amino Acid Metabolism in Liver Diseases
- Lin Yu 1,†,
- Lu Zhang 1,†,
- Xiaoqi Sun 2,*,
- Jingjing Liu 3,*,
- Min Wu 4,*
Author Information
Received: 30 Sep 2025 | Revised: 05 Jan 2026 | Accepted: 13 Jan 2026 | Published: 23 Jan 2026
Abstract
Graphical Abstract
References
- 1.
Huang, D.Q.; Wong, V.W.S.; Rinella, M.E.; et al. Metabolic dysfunction-associated steatotic liver disease in adults. Nat. Rev. Dis. Primers 2025, 11, 14. https://doi.org/10.1038/s41572-025-00599-1.
- 2.
Riazi, K.; Azhari, H.; Charette, J.H.; et al. The prevalence and incidence of NAFLD worldwide: A systematic review and meta-analysis. Lancet Gastroenterol. Hepatol. 2022, 7, 851–861. https://doi.org/10.1016/s2468-1253(22)00165-0.
- 3.
Sheka, A.C.; Adeyi, O.; Thompson, J.; et al. Nonalcoholic Steatohepatitis: A Review. JAMA 2020, 323, 1175–1183. https://doi.org/10.1001/jama.2020.2298.
- 4.
Chalasani, N.; Younossi, Z.; Lavine, J.E.; et al. The diagnosis and management of nonalcoholic fatty liver disease: Practice guidance from the American Association for the Study of Liver Diseases. Hepatology 2018, 67, 328–357. https://doi.org/10.1002/hep.29367.
- 5.
Loomba, R.; Friedman, S.L.; Shulman, G.I. Mechanisms and disease consequences of nonalcoholic fatty liver disease. Cell 2021, 184, 2537–2564. https://doi.org/10.1016/j.cell.2021.04.015.
- 6.
Wang, X.; Zhang, L.; Dong, B. Molecular mechanisms in MASLD/MASH-related HCC. Hepatology 2025, 82, 1303–1324. https://doi.org/10.1097/hep.0000000000000786.
- 7.
Liao, Y.; Chen, Q.; Liu, L.; et al. Amino acid is a major carbon source for hepatic lipogenesis. Cell Metab. 2024, 36, 2437–2448.e8. https://doi.org/10.1016/j.cmet.2024.10.001.
- 8.
Wang, H.; Lun, Y.; Xu, D.; et al. Research progress and therapeutic strategies in hepatocellular carcinoma metabolic reprogramming. J. Adv. Res. 2025, in press. https://doi.org/10.1016/j.jare.2025.08.023.
- 9.
Villar, V.H.; Allega, M.F.; Deshmukh, R.; et al. Hepatic glutamine synthetase controls N5-methylglutamine in homeostasis and cancer. Nat. Chem. Biol. 2023, 19, 292–300. https://doi.org/10.1038/s41589-022-01154-9.
- 10.
Wei, X.; Mo, X.; An, F.; et al. 2′,4′-Dihydroxy-6′-methoxy-3′,5′-dimethylchalcone, a potent Nrf2/ARE pathway inhibitor, reverses drug resistance by decreasing glutathione synthesis and drug efflux in BEL-7402/5-FU cells. Food Chem. Toxicol. 2018, 119, 252–259. https://doi.org/10.1016/j.fct.2018.04.001.
- 11.
Dong, L.; Lou, W.; Xu, C.; et al. Naringenin cationic lipid-modified nanoparticles mitigate MASLD progression by modulating lipid homeostasis and gut microbiota. J. Nanobiotechnol. 2025, 23, 168. https://doi.org/10.1186/s12951-025-03228-x.
- 12.
Wang, M.; Liu, Y.; Li, Y.; et al. Tumor Microenvironment-Responsive Nanoparticles Enhance IDO1 Blockade Immunotherapy by Remodeling Metabolic Immunosuppression. Adv. Sci. 2025, 12, 2405845. https://doi.org/10.1002/advs.202405845.
- 13.
Xiao, F.; Guo, F. Impacts of essential amino acids on energy balance. Mol. Metab. 2022, 57, 101393. https://doi.org/10.1016/j.molmet.2021.101393.
- 14.
Shimomura, Y.; Murakami, T.; Nakai, N.; et al. Exercise promotes BCAA catabolism: Effects of BCAA supplementation on skeletal muscle during exercise. J. Nutr. 2004, 134, 1583s–1587s. https://doi.org/10.1093/jn/134.6.1583S.
- 15.
Dimou, A.; Tsimihodimos, V.; Bairaktari, E. The Critical Role of the Branched Chain Amino Acids (BCAAs) Catabolism-Regulating Enzymes, Branched-Chain Aminotransferase (BCAT) and Branched-Chain α-Keto Acid Dehydrogenase (BCKD), in Human Pathophysiology. Int. J. Mol. Sci. 2022, 23, 4022. https://doi.org/10.3390/ijms23074022.
- 16.
Wegermann, K.; Henao, R.; Diehl, A.M.; et al. Branched chain amino acid transaminase 1 (BCAT1) is overexpressed and hypomethylated in patients with non-alcoholic fatty liver disease who experience adverse clinical events: A pilot study. PLoS ONE 2018, 13, e0204308. https://doi.org/10.1371/journal.pone.0204308.
- 17.
Suryawan, A.; Hawes, J.W.; Harris, R.A.; et al. A molecular model of human branched-chain amino acid metabolism. Am. J. Clin. Nutr. 1998, 68, 72–81. https://doi.org/10.1093/ajcn/68.1.72.
- 18.
Cheng, S.; Wiklund, P.; Autio, R.; et al. Adipose Tissue Dysfunction and Altered Systemic Amino Acid Metabolism Are Associated with Non-Alcoholic Fatty Liver Disease. PLoS ONE 2015, 10, e0138889. https://doi.org/10.1371/journal.pone.0138889.
- 19.
Galarregui, C.; Cantero, I.; Marin-Alejandre, B.A.; et al. Dietary intake of specific amino acids and liver status in subjects with nonalcoholic fatty liver disease: Fatty liver in obesity (FLiO) study. Eur. J. Nutr. 2021, 60, 1769–1780. https://doi.org/10.1007/s00394-020-02370-6.
- 20.
Jian, H.; Li, R.; Huang, X.; et al. Branched-chain amino acids alleviate NAFLD via inhibiting de novo lipogenesis and activating fatty acid β-oxidation in laying hens. Redox Biol. 2024, 77, 103385. https://doi.org/10.1016/j.redox.2024.103385.
- 21.
Felicianna; Lo, E.K.K.; Chen, C.; et al. Low-dose valine attenuates diet-induced metabolic dysfunction-associated steatotic liver disease (MASLD) in mice by enhancing leptin sensitivity and modulating the gut microbiome. Mol. Metab. 2024, 90, 102059. https://doi.org/10.1016/j.molmet.2024.102059.
- 22.
Yu, D.; Richardson, N.E.; Green, C.L.; et al. The adverse metabolic effects of branched-chain amino acids are mediated by isoleucine and valine. Cell Metab. 2021, 33, 905–922.e906. https://doi.org/10.1016/j.cmet.2021.03.025.
- 23.
Zhang, Y.; Lin, S.; Peng, J.; et al. Amelioration of hepatic steatosis by dietary essential amino acid-induced ubiquitination. Mol. Cell 2022, 82, 1528–1542.e10. https://doi.org/10.1016/j.molcel.2022.01.021.
- 24.
Solon-Biet, S.M.; Cogger, V.C.; Pulpitel, T.; et al. Branched chain amino acids impact health and lifespan indirectly via amino acid balance and appetite control. Nat. Metab. 2019, 1, 532–545. https://doi.org/10.1038/s42255-019-0059-2.
- 25.
Rom, O.; Liu, Y.; Liu, Z.; et al. Glycine-based treatment ameliorates NAFLD by modulating fatty acid oxidation, glutathione synthesis, and the gut microbiome. Sci. Transl. Med. 2020, 12, eaaz2841. https://doi.org/10.1126/scitranslmed.aaz2841.
- 26.
Chalasani, N.; Vuppalanchi, R.; Rinella, M.; et al. Randomised clinical trial: A leucine-metformin-sildenafil combination (NS-0200) vs. placebo in patients with non-alcoholic fatty liver disease. Aliment. Pharmacol. Ther. 2018, 47, 1639–1651. https://doi.org/10.1111/apt.14674.
- 27.
Shao, M.; Ye, Z.; Qin, Y.; et al. Abnormal metabolic processes involved in the pathogenesis of non-alcoholic fatty liver disease (Review). Exp. Ther. Med. 2020, 20, 26. https://doi.org/10.3892/etm.2020.9154.
- 28.
Jin, R.; Banton, S.; Tran, V.T.; et al. Amino Acid Metabolism is Altered in Adolescents with Nonalcoholic Fatty Liver Disease-An Untargeted, High Resolution Metabolomics Study. J. Pediatr. 2016, 172, 14–19.e15. https://doi.org/10.1016/j.jpeds.2016.01.026.
- 29.
de Mello, V.D.; Sehgal, R.; Männistö, V.; et al. Serum aromatic and branched-chain amino acids associated with NASH demonstrate divergent associations with serum lipids. Liver Int. 2021, 41, 754–763. https://doi.org/10.1111/liv.14743.
- 30.
Hasegawa, T.; Iino, C.; Endo, T.; et al. Changed Amino Acids in NAFLD and Liver Fibrosis: A Large Cross-Sectional Study without Influence of Insulin Resistance. Nutrients 2020, 12, 1450. https://doi.org/10.3390/nu12051450.
- 31.
Sano, A.; Kakazu, E.; Hamada, S.; et al. Steatotic Hepatocytes Release Mature VLDL Through Methionine and Tyrosine Metabolism in a Keap1-Nrf2-Dependent Manner. Hepatology 2021, 74, 1271–1286. https://doi.org/10.1002/hep.31808.
- 32.
Liu, Q.; Li, X.; Pan, Y.; et al. Efficacy and safety of Qushi Huayu, a traditional Chinese medicine, in patients with nonalcoholic fatty liver disease in a randomized controlled trial. Phytomedicine 2024, 130, 155398. https://doi.org/10.1016/j.phymed.2024.155398.
- 33.
Wu, L.; Dong, X.; Sun, W.; et al. Hydroxysafflor yellow A alleviates oxidative stress and inflammatory damage in the livers of mice with nonalcoholic fatty liver disease and modulates gut microbiota. Front. Pharmacol. 2025, 16, 1568608. https://doi.org/10.3389/fphar.2025.1568608.
- 34.
Choi, W.; Namkung, J.; Hwang, I.; et al. Serotonin signals through a gut-liver axis to regulate hepatic steatosis. Nat. Commun. 2018, 9, 4824. https://doi.org/10.1038/s41467-018-07287-7.
- 35.
Kim, M.; Choi, W.; Yoon, J.; et al. Synthesis and biological evaluation of tyrosine derivatives as peripheral 5HT(2A) receptor antagonists for nonalcoholic fatty liver disease. Eur. J. Med. Chem. 2022, 239, 114517. https://doi.org/10.1016/j.ejmech.2022.114517.
- 36.
Luo, Z.; Liu, Y.; Wang, X.; et al. Exploring tryptophan metabolism: The transition from disturbed balance to diagnostic and therapeutic potential in metabolic diseases. Biochem. Pharmacol. 2024, 230, 116554. https://doi.org/10.1016/j.bcp.2024.116554.
- 37.
Celinski, K.; Konturek, P.C.; Slomka, M.; et al. Effects of treatment with melatonin and tryptophan on liver enzymes, parameters of fat metabolism and plasma levels of cytokines in patients with non-alcoholic fatty liver disease--14 months follow up. J. Physiol. Pharmacol. 2014, 65, 75–82.
- 38.
Ritze, Y.; Bárdos, G.; Hubert, A.; et al. Effect of tryptophan supplementation on diet-induced non-alcoholic fatty liver disease in mice. Br. J. Nutr. 2014, 112, 1–7. https://doi.org/10.1017/s0007114514000440.
- 39.
Zhou, Q.; Shi, Y.; Chen, C.; et al. A narrative review of the roles of indoleamine 2,3-dioxygenase and tryptophan-2,3-dioxygenase in liver diseases. Ann. Transl. Med. 2021, 9, 174. https://doi.org/10.21037/atm-20-3594.
- 40.
Pyun, D.H.; Kim, T.J.; Kim, M.J.; et al. Endogenous metabolite, kynurenic acid, attenuates nonalcoholic fatty liver disease via AMPK/autophagy- and AMPK/ORP150-mediated signaling. J. Cell Physiol. 2021, 236, 4902–4912. https://doi.org/10.1002/jcp.30199.
- 41.
Agudelo, L.Z.; Ferreira, D.M.S.; Cervenka, I.; et al. Kynurenic Acid and Gpr35 Regulate Adipose Tissue Energy Homeostasis and Inflammation. Cell Metab. 2018, 27, 378–392.e5. https://doi.org/10.1016/j.cmet.2018.01.004.
- 42.
Nagano, J.; Shimizu, M.; Hara, T.; et al. Effects of indoleamine 2,3-dioxygenase deficiency on high-fat diet-induced hepatic inflammation. PLoS ONE 2013, 8, e73404. https://doi.org/10.1371/journal.pone.0073404.
- 43.
Laurans, L.; Venteclef, N.; Haddad, Y.; et al. Genetic deficiency of indoleamine 2,3-dioxygenase promotes gut microbiota-mediated metabolic health. Nat. Med. 2018, 24, 1113–1120. https://doi.org/10.1038/s41591-018-0060-4.
- 44.
Zhu, Y.; Shang, L.; Tang, Y.; et al. Genome-Wide Profiling of H3K27ac Identifies TDO2 as a Pivotal Therapeutic Target in Metabolic Associated Steatohepatitis Liver Disease. Adv. Sci. 2024, 11, e2404224. https://doi.org/10.1002/advs.202404224.
- 45.
Nonogaki, K.; Kaji, T. Whey protein isolate inhibits hepatic FGF21 production, which precedes weight gain, hyperinsulinemia and hyperglycemia in mice fed a high-fat diet. Sci. Rep. 2020, 10, 15784. https://doi.org/10.1038/s41598-020-72975-8.
- 46.
Zhang, K.; Li, X.; Wang, X.; et al. Gut Barrier Proteins Mediate Liver Regulation by the Effects of Serotonin on the Non-Alcoholic Fatty Liver Disease. Curr. Protein Pept. Sci. 2020, 21, 978–984. https://doi.org/10.2174/1389203721666200615171928.
- 47.
Gao, Y.; Chen, Q.; Yang, S.; et al. Indole alleviates nonalcoholic fatty liver disease in an ACE2-dependent manner. Faseb J. 2024, 38, e70061. https://doi.org/10.1096/fj.202401172RR.
- 48.
Min, B.H.; Devi, S.; Kwon, G.H.; et al. Gut microbiota-derived indole compounds attenuate metabolic dysfunction-associated steatotic liver disease by improving fat metabolism and inflammation. Gut Microbes 2024, 16, 2307568. https://doi.org/10.1080/19490976.2024.2307568.
- 49.
Zhang, C.; Fu, Q.; Shao, K.; et al. Indole-3-acetic acid improves the hepatic mitochondrial respiration defects by PGC1a up-regulation. Cell Signal 2022, 99, 110442. https://doi.org/10.1016/j.cellsig.2022.110442.
- 50.
Zhao, Z.H.; Xin, F.Z.; Xue, Y.; et al. Indole-3-propionic acid inhibits gut dysbiosis and endotoxin leakage to attenuate steatohepatitis in rats. Exp. Mol. Med. 2019, 51, 1–14. https://doi.org/10.1038/s12276-019-0304-5.
- 51.
Deng, Y.; Hu, M.; Huang, S.; et al. Molecular mechanism and therapeutic significance of essential amino acids in metabolically associated fatty liver disease. J. Nutr. Biochem. 2024, 126, 109581. https://doi.org/10.1016/j.jnutbio.2024.109581.
- 52.
Aron-Wisnewsky, J.; Prifti, E.; Belda, E.; et al. Major microbiota dysbiosis in severe obesity: Fate after bariatric surgery. Gut 2019, 68, 70–82. https://doi.org/10.1136/gutjnl-2018-316103.
- 53.
Feng, R.N.; Niu, Y.C.; Sun, X.W.; et al. Histidine supplementation improves insulin resistance through suppressed inflammation in obese women with the metabolic syndrome: A randomised controlled trial. Diabetologia 2013, 56, 985–994. https://doi.org/10.1007/s00125-013-2839-7.
- 54.
Koh, A.; Molinaro, A.; Ståhlman, M.; et al. Microbially Produced Imidazole Propionate Impairs Insulin Signaling through mTORC1. Cell 2018, 175, 947–961.e17. https://doi.org/10.1016/j.cell.2018.09.055.
- 55.
Molinaro, A.; Bel Lassen, P.; Henricsson, M.; et al. Imidazole propionate is increased in diabetes and associated with dietary patterns and altered microbial ecology. Nat. Commun. 2020, 11, 5881. https://doi.org/10.1038/s41467-020-19589-w.
- 56.
Quesada-Vázquez, S.; Castells-Nobau, A.; Latorre, J.; et al. Potential therapeutic implications of histidine catabolism by the gut microbiota in NAFLD patients with morbid obesity. Cell Rep. Med. 2023, 4, 101341. https://doi.org/10.1016/j.xcrm.2023.101341.
- 57.
Fujimi, T.J.; Sate, M.; Tsuchiya, M.; et al. Gene Expression and Histochemical Analyses in the Fatty Livers of Rats Fed a Histidine-Excess Diet. J. Nutr. Sci. Vitaminol. 2020, 66, 561–570. https://doi.org/10.3177/jnsv.66.561.
- 58.
Kennedy, L.; Hargrove, L.; Demieville, J.; et al. Knockout of l-Histidine Decarboxylase Prevents Cholangiocyte Damage and Hepatic Fibrosis in Mice Subjected to High-Fat Diet Feeding via Disrupted Histamine/Leptin Signaling. Am. J. Pathol. 2018, 188, 600–615. https://doi.org/10.1016/j.ajpath.2017.11.016.
- 59.
Yamada, S.; Tanimoto, A.; Sasaguri, Y. Critical in vivo roles of histamine and histamine receptor signaling in animal models of metabolic syndrome. Pathol. Int. 2016, 66, 661–671. https://doi.org/10.1111/pin.12477.
- 60.
Martínez-Chantar, M.L.; García-Trevijano, E.R.; Latasa, M.U.; et al. Methionine adenosyltransferase II beta subunit gene expression provides a proliferative advantage in human hepatoma. Gastroenterology 2003, 124, 940–948. https://doi.org/10.1053/gast.2003.50151.
- 61.
Pacana, T.; Cazanave, S.; Verdianelli, A.; et al. Dysregulated Hepatic Methionine Metabolism Drives Homocysteine Elevation in Diet-Induced Nonalcoholic Fatty Liver Disease. PLoS ONE 2015, 10, e0136822. https://doi.org/10.1371/journal.pone.0136822.
- 62.
Yun, K.U.; Ryu, C.S.; Oh, J.M.; et al. Plasma homocysteine level and hepatic sulfur amino acid metabolism in mice fed a high-fat diet. Eur. J. Nutr. 2013, 52, 127–134. https://doi.org/10.1007/s00394-011-0294-0.
- 63.
Kwon, D.Y.; Jung, Y.S.; Kim, S.J.; et al. Impaired sulfur-amino acid metabolism and oxidative stress in nonalcoholic fatty liver are alleviated by betaine supplementation in rats. J. Nutr. 2009, 139, 63–68. https://doi.org/10.3945/jn.108.094771.
- 64.
Fling, R.R.; Doskey, C.M.; Fader, K.A.; et al. 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) dysregulates hepatic one carbon metabolism during the progression of steatosis to steatohepatitis with fibrosis in mice. Sci. Rep. 2020, 10, 14831. https://doi.org/10.1038/s41598-020-71795-0.
- 65.
Ramani, K.; Yang, H.; Kuhlenkamp, J.; et al. Changes in the expression of methionine adenosyltransferase genes and S-adenosylmethionine homeostasis during hepatic stellate cell activation. Hepatology 2010, 51, 986–995. https://doi.org/10.1002/hep.23411.
- 66.
Yamakado, M.; Tanaka, T.; Nagao, K.; et al. Plasma amino acid profile associated with fatty liver disease and co-occurrence of metabolic risk factors. Sci. Rep. 2017, 7, 14485. https://doi.org/10.1038/s41598-017-14974-w.
- 67.
Navik, U.; Sheth, V.G.; Sharma, N.; et al. L-Methionine supplementation attenuates high-fat fructose diet-induced non-alcoholic steatohepatitis by modulating lipid metabolism, fibrosis, and inflammation in rats. Food Funct. 2022, 13, 4941–4953. https://doi.org/10.1039/d1fo03403k.
- 68.
Curcio, A.; Romano, A.; Cuozzo, S.; et al. Silymarin in Combination with Vitamin C, Vitamin E, Coenzyme Q10 and Selenomethionine to Improve Liver Enzymes and Blood Lipid Profile in NAFLD Patients. Medicina 2020, 56, 544. https://doi.org/10.3390/medicina56100544.
- 69.
Aissa, A.F.; Tryndyak, V.; de Conti, A.; et al. Effect of methionine-deficient and methionine-supplemented diets on the hepatic one-carbon and lipid metabolism in mice. Mol. Nutr. Food Res. 2014, 58, 1502–1512. https://doi.org/10.1002/mnfr.201300726.
- 70.
Martínez-Uña, M.; Varela-Rey, M.; Cano, A.; et al. Excess S-adenosylmethionine reroutes phosphatidylethanolamine towards phosphatidylcholine and triglyceride synthesis. Hepatology 2013, 58, 1296–1305. https://doi.org/10.1002/hep.26399.
- 71.
Yamada, H.; Akahoshi, N.; Kamata, S.; et al. Methionine excess in diet induces acute lethal hepatitis in mice lacking cystathionine γ-lyase, an animal model of cystathioninuria. Free Radic. Biol. Med. 2012, 52, 1716–1726. https://doi.org/10.1016/j.freeradbiomed.2012.02.033.
- 72.
Robinson, A.E.; Binek, A.; Ramani, K.; et al. Hyperphosphorylation of hepatic proteome characterizes nonalcoholic fatty liver disease in S-adenosylmethionine deficiency. iScience 2023, 26, 105987. https://doi.org/10.1016/j.isci.2023.105987.
- 73.
Walkey, C.J.; Donohue, L.R.; Bronson, R.; et al. Disruption of the murine gene encoding phosphatidylethanolamine N-methyltransferase. Proc. Natl. Acad. Sci. USA 1997, 94, 12880–12885. https://doi.org/10.1073/pnas.94.24.12880.
- 74.
van der Veen, J.N.; Kennelly, J.P.; Wan, S.; et al. The critical role of phosphatidylcholine and phosphatidylethanolamine metabolism in health and disease. Biochim. Biophys. Acta Biomembr. 2017, 1859, 1558–1572. https://doi.org/10.1016/j.bbamem.2017.04.006.
- 75.
Luka, Z.; Capdevila, A.; Mato, J.M.; et al. A glycine N-methyltransferase knockout mouse model for humans with deficiency of this enzyme. Transgenic Res. 2006, 15, 393–397. https://doi.org/10.1007/s11248-006-0008-1.
- 76.
Martínez-Chantar, M.L.; Vázquez-Chantada, M.; Ariz, U.; et al. Loss of the glycine N-methyltransferase gene leads to steatosis and hepatocellular carcinoma in mice. Hepatology 2008, 47, 1191–1199. https://doi.org/10.1002/hep.22159.
- 77.
Choumenkovitch, S.F.; Selhub, J.; Bagley, P.J.; et al. In the cystathionine beta-synthase knockout mouse, elevations in total plasma homocysteine increase tissue S-adenosylhomocysteine, but responses of S-adenosylmethionine and DNA methylation are tissue specific. J. Nutr. 2002, 132, 2157–2160. https://doi.org/10.1093/jn/132.8.2157.
- 78.
Kruger, W.D. Cystathionine β-synthase deficiency: Of mice and men. Mol. Genet. Metab. 2017, 121, 199–205. https://doi.org/10.1016/j.ymgme.2017.05.011.
- 79.
Jacobs, R.L.; Jiang, H.; Kennelly, J.P.; et al. Cystathionine beta-synthase deficiency alters hepatic phospholipid and choline metabolism: Post-translational repression of phosphatidylethanolamine N-methyltransferase is a consequence rather than a cause of liver injury in homocystinuria. Mol. Genet. Metab. 2017, 120, 325–336. https://doi.org/10.1016/j.ymgme.2017.02.010.
- 80.
Teng, Y.W.; Mehedint, M.G.; Garrow, T.A.; et al. Deletion of betaine-homocysteine S-methyltransferase in mice perturbs choline and 1-carbon metabolism, resulting in fatty liver and hepatocellular carcinomas. J. Biol. Chem. 2011, 286, 36258–36267. https://doi.org/10.1074/jbc.M111.265348.
- 81.
Li, Z.; Wang, F.; Liang, B.; et al. Methionine metabolism in chronic liver diseases: An update on molecular mechanism and therapeutic implication. Signal Transduct. Target. Ther. 2020, 5, 280. https://doi.org/10.1038/s41392-020-00349-7.
- 82.
da Silva, R.P.; Kelly, K.B.; Al Rajabi, A.; et al. Novel insights on interactions between folate and lipid metabolism. Biofactors 2014, 40, 277–283. https://doi.org/10.1002/biof.1154.
- 83.
Christensen, K.E.; Wu, Q.; Wang, X.; et al. Steatosis in mice is associated with gender, folate intake, and expression of genes of one-carbon metabolism. J. Nutr. 2010, 140, 1736–1741. https://doi.org/10.3945/jn.110.124917.
- 84.
Champier, J.; Claustrat, F.; Nazaret, N.; et al. Folate depletion changes gene expression of fatty acid metabolism, DNA synthesis, and circadian cycle in male mice. Nutr. Res. 2012, 32, 124–132. https://doi.org/10.1016/j.nutres.2011.12.012.
- 85.
Polyzos, S.A.; Kountouras, J.; Patsiaoura, K.; et al. Serum homocysteine levels in patients with nonalcoholic fatty liver disease. Ann. Hepatol. 2012, 11, 68–76.
- 86.
Koplay, M.; Gulcan, E.; Ozkan, F. Association between serum vitamin B12 levels and the degree of steatosis in patients with nonalcoholic fatty liver disease. J. Investig. Med. 2011, 59, 1137–1140. https://doi.org/10.2310/JIM.0b013e31822a29f5.
- 87.
Mahamid, M.; Mahroum, N.; Bragazzi, N.L.; et al. Folate and B12 Levels Correlate with Histological Severity in NASH Patients. Nutrients 2018, 10, 440. https://doi.org/10.3390/nu10040440.
- 88.
Vahedi, H.; Bavafaetousi, N.; Zolfaghari, P.; et al. Association between serum folate levels and fatty liver disease. Clin. Nutr. Exp. 2020, 29, 30–35. https://doi.org/10.1016/j.yclnex.2019.11.004.
- 89.
Tripathi, M.; Singh, B.K.; Zhou, J.; et al. Vitamin B12 and folate decrease inflammation and fibrosis in NASH by preventing syntaxin 17 homocysteinylation. J. Hepatol. 2022, 77, 1246–1255. https://doi.org/10.1016/j.jhep.2022.06.033.
- 90.
McBride, M.J.; Hunter, C.J.; Zhang, Z.; et al. Glycine homeostasis requires reverse SHMT flux. Cell Metab. 2024, 36, 103–115.e104. https://doi.org/10.1016/j.cmet.2023.12.001.
- 91.
Newgard, C.B.; An, J.; Bain, J.R.; et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009, 9, 311–326. https://doi.org/10.1016/j.cmet.2009.02.002.
- 92.
Gaggini, M.; Carli, F.; Rosso, C.; et al. Altered amino acid concentrations in NAFLD: Impact of obesity and insulin resistance. Hepatology 2018, 67, 145–158. https://doi.org/10.1002/hep.29465.
- 93.
Ghrayeb, A.; Finney, A.C.; Agranovich, B.; et al. Serine synthesis via reversed SHMT2 activity drives glycine depletion and acetaminophen hepatotoxicity in MASLD. Cell Metab. 2024, 36, 116–129.e117. https://doi.org/10.1016/j.cmet.2023.12.013.
- 94.
Chen, G.; Zhou, G.; Zhai, L.; et al. SHMT2 reduces fatty liver but is necessary for liver inflammation and fibrosis in mice. Commun. Biol. 2024, 7, 173. https://doi.org/10.1038/s42003-024-05861-y.
- 95.
Mardinoglu, A.; Agren, R.; Kampf, C.; et al. Genome-scale metabolic modelling of hepatocytes reveals serine deficiency in patients with non-alcoholic fatty liver disease. Nat. Commun. 2014, 5, 3083. https://doi.org/10.1038/ncomms4083.
- 96.
Holm, L.J.; Haupt-Jorgensen, M.; Larsen, J.; et al. L-serine supplementation lowers diabetes incidence and improves blood glucose homeostasis in NOD mice. PLoS ONE 2018, 13, e0194414. https://doi.org/10.1371/journal.pone.0194414.
- 97.
Chen, H.; Liu, C.; Wang, Q.; et al. Renal UTX-PHGDH-serine axis regulates metabolic disorders in the kidney and liver. Nat. Commun. 2022, 13, 3835. https://doi.org/10.1038/s41467-022-31476-0.
- 98.
Mino, M.; Kakazu, E.; Sano, A.; et al. Comprehensive analysis of peripheral blood free amino acids in MASLD: The impact of glycine-serine-threonine metabolism. Amino Acids 2024, 57, 3. https://doi.org/10.1007/s00726-024-03433-2.
- 99.
Wei, Y.; Wang, Y.G.; Jia, Y.; et al. Liver homeostasis is maintained by midlobular zone 2 hepatocytes. Science 2021, 371, eabb1625. https://doi.org/10.1126/science.abb1625.
- 100.
Hakvoort, T.B.; He, Y.; Kulik, W.; et al. Pivotal role of glutamine synthetase in ammonia detoxification. Hepatology 2017, 65, 281–293. https://doi.org/10.1002/hep.28852.
- 101.
Gebhardt, R.; Mecke, D. Heterogeneous distribution of glutamine synthetase among rat liver parenchymal cells in situ and in primary culture. Embo J. 1983, 2, 567–570. https://doi.org/10.1002/j.1460-2075.1983.tb01464.x.
- 102.
Zhao, J.; Zeng, J.; Zhu, C.; et al. Genetically predicted plasma levels of amino acids and metabolic dysfunction-associated fatty liver disease risk: A Mendelian randomization study. BMC Med. 2023, 21, 469. https://doi.org/10.1186/s12916-023-03185-y.
- 103.
Zhou, X.; Zhang, J.; Sun, Y.; et al. Glutamine Ameliorates Liver Steatosis via Regulation of Glycolipid Metabolism and Gut Microbiota in High-Fat Diet-Induced Obese Mice. J. Agric. Food Chem. 2023, 71, 15656–15667. https://doi.org/10.1021/acs.jafc.3c05566.
- 104.
Leite, J.S.M.; Vilas-Boas, E.A.; Takahashi, H.K.; et al. Liver lipid metabolism, oxidative stress, and inflammation in glutamine-supplemented ob/ob mice. J. Nutr. Biochem. 2025, 138, 109842. https://doi.org/10.1016/j.jnutbio.2025.109842.
- 105.
Yan, R.; Cai, H.; Zhou, X.; et al. Hypoxia-inducible factor-2α promotes fibrosis in non-alcoholic fatty liver disease by enhancing glutamine catabolism and inhibiting yes-associated protein phosphorylation in hepatic stellate cells. Front. Endocrinol. 2024, 15, 1344971. https://doi.org/10.3389/fendo.2024.1344971.
- 106.
Du, K.; Chitneni, S.K.; Suzuki, A.; et al. Increased Glutaminolysis Marks Active Scarring in Nonalcoholic Steatohepatitis Progression. Cell Mol. Gastroenterol. Hepatol. 2020, 10, 1–21. https://doi.org/10.1016/j.jcmgh.2019.12.006.
- 107.
Simon, J.; Nuñez-García, M.; Fernández-Tussy, P.; et al. Targeting Hepatic Glutaminase 1 Ameliorates Non-alcoholic Steatohepatitis by Restoring Very-Low-Density Lipoprotein Triglyceride Assembly. Cell Metab. 2020, 31, 605–622.e610. https://doi.org/10.1016/j.cmet.2020.01.013.
- 108.
Shen, J.; Xie, E.; Shen, S.; et al. Essentiality of SLC7A11-mediated nonessential amino acids in MASLD. Sci. Bull. 2024, 69, 3700–3716. https://doi.org/10.1016/j.scib.2024.09.019.
- 109.
Park, S.; Hall, M.N. Metabolic reprogramming in hepatocellular carcinoma: Mechanisms and therapeutic implications. Exp. Mol. Med. 2025, 57, 515–523. https://doi.org/10.1038/s12276-025-01415-2.
- 110.
Vettore, L.; Westbrook, R.L.; Tennant, D.A. New aspects of amino acid metabolism in cancer. Br. J. Cancer 2020, 122, 150–156. https://doi.org/10.1038/s41416-019-0620-5.
- 111.
Wang, N.; Lu, S.; Cao, Z.; et al. Pyruvate metabolism enzyme DLAT promotes tumorigenesis by suppressing leucine catabolism. Cell Metab. 2025, 37, 1381–1399.e9. https://doi.org/10.1016/j.cmet.2025.02.008.
- 112.
Watanabe, A.; Higashi, T.; Sakata, T.; et al. Serum Amino Acid Levels in Patients With Hepatocellular Carcinoma. Cancer 1984, 54, 1875–1882. https://doi.org/10.1002/1097-0142(19841101)54:9<1875::AID-CNCR2820540918>3.0.CO;2-O.
- 113.
Cai, D.; Ji, J.; Yang, C.; et al. Branched-Chain Amino Acid Metabolic Reprogramming and Cancer: Molecular Mechanisms, Immune Regulation, and Precision Targeting. Oncol. Res. 2025, 34, 9.
- 114.
Wu, T.; Zheng, X.; Yang, M.; et al. Serum Amino Acid Profiles Predict the Development of Hepatocellular Carcinoma in Patients with Chronic HBV Infection. ACS Omega 2022, 7, 15795–15808. https://doi.org/10.1021/acsomega.2c00885.
- 115.
Ericksen, R.; Lim, S.L.; McDonnell, E.; et al. Loss of BCAA Catabolism during Carcinogenesis Enhances mTORC1 Activity and Promotes Tumor Development and Progression. Cell Metab. 2019, 29, 1151–1165. https://doi.org/10.1016/j.cmet.2018.12.020.
- 116.
Liu, Y.; Wang, F.; Yan, G.; et al. CPT1A loss disrupts BCAA metabolism to confer therapeutic vulnerability in TP53-mutated liver cancer. Cancer Lett. 2024, 595, 217006. https://doi.org/10.1016/j.canlet.2024.217006.
- 117.
Qian, L.; Li, N.; Lu, X.C.; et al. Enhanced BCAT1 activity and BCAA metabolism promotes RhoC activity in cancer progression. Nat. Metab. 2023, 5, 1159–1173. https://doi.org/10.1038/s42255-023-00818-7.
- 118.
Yang, D.; Liu, H.; Cai, Y.; et al. Branched-chain amino acid catabolism breaks glutamine addiction to sustain hepatocellular carcinoma progression. Cell Rep. 2022, 41, 111691. https://doi.org/10.1016/j.celrep.2022.111691.
- 119.
Xue, C.; Li, G.; Zheng, Q.; et al. Tryptophan metabolism in health and disease. Cell Metab. 2023, 35, 1304–1326.
- 120.
Seo, S.-K.; Kwon, B. Immune regulation through tryptophan metabolism. Exp. Mol. Med. 2023, 55, 1371–1379. https://doi.org/10.1038/s12276-023-01028-7.
- 121.
Savitz, J. The kynurenine pathway: A finger in every pie. Mol. Psychiatry 2020, 25, 131–147.
- 122.
Stepien, M.; Duarte-Salles, T.; Fedirko, V.; et al. Alteration of amino acid and biogenic amine metabolism in hepatobiliary cancers: Findings from a prospective cohort study. Int. J. Cancer 2016, 138, 348–360. https://doi.org/10.1002/ijc.29718.
- 123.
Bekki, S.; Hashimoto, S.; Yamasaki, K.; et al. Serum kynurenine levels are a novel biomarker to predict the prognosis of patients with hepatocellular carcinoma. PLoS ONE 2020, 15, e0241002. https://doi.org/10.1371/journal.pone.0241002.
- 124.
Tang, Z.; Bai, Y.; Fang, Q.; et al. Spatial transcriptomics reveals tryptophan metabolism restricting maturation of intratumoral tertiary lymphoid structures. Cancer Cell 2025, 43, 1025–1044.e1014. https://doi.org/10.1016/j.ccell.2025.03.011.
- 125.
Dey, A.; Jones, J.E.; Nebert, D.W. Tissue- and cell type-specific expression of cytochrome P450 1A1 and cytochrome P450 1A2 mRNA in the mouse localized in situ hybridization. Biochem. Pharmacol. 1999, 58, 525–537. https://doi.org/10.1016/S0006-2952(99)00110-0.
- 126.
Simile, M.M.; Cigliano, A.; Paliogiannis, P.; et al. Nuclear localization dictates hepatocarcinogenesis suppression by glycine N-methyltransferase. Transl. Oncol. 2022, 15, 101239. https://doi.org/10.1016/j.tranon.2021.101239.
- 127.
Liu, P.; Zhou, Y.; Dong, X.; et al. ZNF165 Is Involved in the Regulation of Immune Microenvironment and Promoting the Proliferation and Migration of Hepatocellular Carcinoma by AhR/CYP1A1. J. Immunol. Res. 2022, 2022, 4446805. https://doi.org/10.1155/2022/4446805.
- 128.
Jin, H.; Zhang, Y.; You, H.; et al. Prognostic significance of kynurenine 3-monooxygenase and effects on proliferation, migration, and invasion of human hepatocellular carcinoma. Sci. Rep. 2015, 5, 10466. https://doi.org/10.1038/srep10466.
- 129.
Xue, C.; Gu, X.; Zheng, Q.; et al. Effects of 3-HAA on HCC by Regulating the Heterogeneous Macrophages—A scRNA-Seq Analysis. Adv. Sci. 2023, 10, 2207074. https://doi.org/10.1002/advs.202207074.
- 130.
Shi, Z.; Gan, G.; Gao, X.; et al. Kynurenine catabolic enzyme KMO regulates HCC growth. Clin. Transl. Med. 2022, 12, e697. https://doi.org/10.1002/ctm2.697.
- 131.
Tummala, K.S.; Gomes, A.L.; Yilmaz, M.; et al. Inhibition of De Novo NAD<SUP>+</SUP> Synthesis by Oncogenic URI Causes Liver Tumorigenesis through DNA Damage. Cancer Cell 2014, 26, 826–839. https://doi.org/10.1016/j.ccell.2014.10.002.
- 132.
Maffei, M.E. 5-Hydroxytryptophan (5-HTP): Natural Occurrence, Analysis, Biosynthesis, Biotechnology, Physiology and Toxicology. Int. J. Mol. Sci. 2020, 22, 181. https://doi.org/10.3390/ijms22010181.
- 133.
Zhu, Y.; Yin, L.; Liu, Q.; et al. Tryptophan metabolic pathway plays a key role in the stress-induced emotional eating. Curr. Res. Food Sci. 2024, 8, 100754. https://doi.org/10.1016/j.crfs.2024.100754.
- 134.
Wang, L.; Deng, Y.; Gao, J.; et al. Biosynthesis of melatonin from l-tryptophan by an engineered microbial cell factory. Biotechnol. Biofuels Bioprod. 2024, 17, 27. https://doi.org/10.1186/s13068-024-02476-7.
- 135.
Dong, R.; Wang, T.; Dong, W.; et al. TGM2-mediated histone serotonylation promotes HCC progression via MYC signalling pathway. J. Hepatol. 2025, 83, 105–118. https://doi.org/10.1016/j.jhep.2024.12.038.
- 136.
Fatima, S.; Shi, X.; Lin, Z.; et al. 5-Hydroxytryptamine promotes hepatocellular carcinoma proliferation by influencing β-catenin. Mol. Oncol. 2016, 10, 195–212. https://doi.org/10.1016/j.molonc.2015.09.008.
- 137.
Zuo, X.; Chen, Z.; Cai, J.; et al. 5-Hydroxytryptamine Receptor 1D Aggravates Hepatocellular Carcinoma Progression Through FoxO6 in AKT-Dependent and Independent Manners. Hepatology 2019, 69, 2031–2047.
- 138.
Liu, S.; Miao, R.; Zhai, M.; et al. Effects and related mechanisms of serotonin on malignant biological behavior of hepatocellular carcinoma via regulation of Yap. Oncotarget 2017, 8, 47412–47424. https://doi.org/10.18632/oncotarget.17658.
- 139.
Sadik, A.; Somarribas Patterson, L.F.; Öztürk, S.; et al. IL4I1 Is a Metabolic Immune Checkpoint that Activates the AHR and Promotes Tumor Progression. Cell 2020, 182, 1252–1270.e34. https://doi.org/10.1016/j.cell.2020.07.038.
- 140.
Venkateswaran, N.; Garcia, R.; Lafita-Navarro, M.C.; et al. Tryptophan fuels MYC-dependent liver tumorigenesis through indole 3-pyruvate synthesis. Nat. Commun. 2024, 15, 4266. https://doi.org/10.1038/s41467-024-47868-3.
- 141.
Hubbard, T.D.; Murray, I.A.; Perdew, G.H. Indole and Tryptophan Metabolism: Endogenous and Dietary Routes to Ah Receptor Activation. Drug Metab. Dispos. 2015, 43, 1522–1535. https://doi.org/10.1124/dmd.115.064246.
- 142.
Chen, W.; Wen, L.; Bao, Y.; et al. Gut flora disequilibrium promotes the initiation of liver cancer by modulating tryptophan metabolism and up-regulating SREBP2. Proc. Natl. Acad. Sci. USA 2022, 119, e2203894119, doi:doi:10.1073/pnas.2203894119.
- 143.
Diaz, G.A.; Bechter, M.; Cederbaum, S.D. The role and control of arginine levels in arginase 1 deficiency. J. Inherit. Metab. Dis. 2023, 46, 3–14. https://doi.org/10.1002/jimd.12564.
- 144.
Heuser, S.; Li, J.; Pudewell, S.; et al. Biochemistry, pharmacology, and in vivo function of arginases. Pharmacol. Rev. 2024, 77, 100015. https://doi.org/10.1124/pharmrev.124.001271.
- 145.
Mossmann, D.; Müller, C.; Park, S.; et al. Arginine reprograms metabolism in liver cancer via RBM39. Cell 2023, 186, 5068–5083. https://doi.org/10.1016/j.cell.2023.09.011.
- 146.
Thongkum, A.; Wu, C.; Li, Y.-Y.; et al. The Combination of Arginine Deprivation and 5-Fluorouracil Improves Therapeutic Efficacy in Argininosuccinate Synthetase Negative Hepatocellular Carcinoma. Int. J. Mol. Sci. 2017, 18, 1175.
- 147.
Bibi, K.; Fatima, T.; Sohrab, S.; et al. Polymorphic variants of ASS1 gene related to arginine metabolism and the risk of HCC. Protein Pept. Lett. 2023, 30, 587–596.
- 148.
Missiaen, R.; Anderson, N.M.; Kim, L.C.; et al. GCN2 inhibition sensitizes arginine-deprived hepatocellular carcinoma cells to senolytic treatment. Cell Metab. 2022, 34, 1151–1167.e7. https://doi.org/10.1016/j.cmet.2022.06.010.
- 149.
Lowman, X.; Hanse, E.; Yang, Y.; et al. p53 Promotes Cancer Cell Adaptation to Glutamine Deprivation by Upregulating Slc7a3 to Increase Arginine Uptake. Cell Rep. 2019, 26, 3051–3060. https://doi.org/10.1016/j.celrep.2019.02.037.
- 150.
Jung, J.W.; Macalino, S.J.Y.; Cui, M.; et al. Transmembrane 4 L Six Family Member 5 Senses Arginine for mTORC1 Signaling. Cell Metab. 2019, 29, 1306–1319.e7. https://doi.org/10.1016/j.cmet.2019.03.005.
- 151.
Kubo, S.; Tamori, A.; Nishiguchi, S.; et al. Relationship of polyamine metabolism to degree of malignancy of human hepatocellular carcinoma. Oncol. Rep. 1998, 5, 1385–1388. https://doi.org/10.3892/OR.5.6.1385.
- 152.
Tamori, A.; Nishiguchi, S.; Kuroki, T.; et al. Relationship of Ornithine Decarboxylase activity and histological findings in human hepatocellular carcinoma. Hepatology 1994, 20, 1179–1186. https://doi.org/10.1002/hep.1840200512.
- 153.
Liu, Z.Y.; Wu, C.Y.; Wu, R.Q.; et al. Efflux of N1-acetylspermidine from hepatoma fosters macrophage-mediated immune suppression to dampen immunotherapeutic efficacy. Immunity 2025, 58, 1572–1585.e10. https://doi.org/10.1016/j.immuni.2025.05.006.
- 154.
Yoo, H.C.; Yu, Y.C.; Sung, Y.; et al. Glutamine reliance in cell metabolism. Exp. Mol. Med. 2020, 52, 1496–1516. https://doi.org/10.1038/s12276-020-00504-8.
- 155.
Yuan, Q.; Yin, L.; He, J.; et al. Metabolism of asparagine in the physiological state and cancer. Cell Commun. Signal. 2024, 22, 163. https://doi.org/10.1186/s12964-024-01540-x.
- 156.
Holeček, M. Serine Metabolism in Health and Disease and as a Conditionally Essential Amino Acid. Nutrients 2022, 14, 1987. https://doi.org/10.3390/nu14091987.
- 157.
Chakrapani, A.; Gissen, P.; McKiernan, P. Disorders of tyrosine metabolism. In Inborn Metabolic Diseases: Diagnosis and Treatment; Springer: Berlin/Heidelberg, Germany, 2022; pp. 355–367.
- 158.
Yang, W.; Zhu, G.; Zhou, G.; et al. Alterations of glutamine and glutamate levels in patients and rats with hepatocellular carcinoma. J. Liq. Chromatogr. Relat. Technol. 2018, 41, 588–594. https://doi.org/10.1080/10826076.2018.1485034.
- 159.
Zhang, Q.; Wei, T.; Jin, W.; et al. Deficiency in SLC25A15, a hypoxia-responsive gene, promotes hepatocellular carcinoma by reprogramming glutamine metabolism. J. Hepatol. 2024, 80, 293–308. https://doi.org/10.1016/j.jhep.2023.10.024.
- 160.
Yu, D.; Shi, X.; Meng, G.; et al. Kidney-type glutaminase (GLS1) is a biomarker for pathologic diagnosis and prognosis of hepatocellular carcinoma. Oncotarget 2015, 6, 7619–7631. https://doi.org/10.18632/oncotarget.3196.
- 161.
Dong, M.; Miao, L.; Zhang, F.; et al. Nuclear factor-κB p65 regulates glutaminase 1 expression in human hepatocellular carcinoma. OncoTargets Ther. 2018, 11, 3721–3729. https://doi.org/10.2147/OTT.S167408.
- 162.
Kaelin, W.G., Jr.; Ratcliffe, P.J. Oxygen Sensing by Metazoans: The Central Role of the HIF Hydroxylase Pathway. Mol. Cell 2008, 30, 393–402. https://doi.org/10.1016/j.molcel.2008.04.009.
- 163.
Selak, M.A.; Armour, S.M.; MacKenzie, E.D.; et al. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-α prolyl hydroxylase. Cancer Cell 2005, 7, 77–85. https://doi.org/10.1016/j.ccr.2004.11.022.
- 164.
Zhang, T.; Cui, Y.; Wu, Y.-Y.; et al. Mitochondrial GCN5L1 regulates glutaminase acetylation and hepatocellular carcinoma. Clin. Transl. Med. 2022, 12, e852. https://doi.org/10.1002/ctm2.852.
- 165.
Cui, Z.; Li, C.; Liu, W.; et al. Scutellarin activates IDH1 to exert antitumor effects in hepatocellular carcinoma progression. Cell Death Dis. 2024, 15, 267. https://doi.org/10.1038/s41419-024-06625-6.
- 166.
Luo, H.; Wang, Q.; Yang, F.; et al. Signaling metabolite succinylacetone activates HIF-1α and promotes angiogenesis in GSTZ1-deficient hepatocellular carcinoma. JCI Insight 2023, 8, e164968. https://doi.org/10.1172/jci.insight.164968.
- 167.
Fedotcheva, N.; Sokolov, A.; Kondrashova, M. Nonezymatic formation of succinate in mitochondria under oxidative stress. Free Radic. Biol. Med. 2006, 41, 56–64. https://doi.org/10.1016/J.FREERADBIOMED.2006.02.012.
- 168.
Guo, W.; Zhao, Y.; Zhang, Z.; et al. Disruption of xCT inhibits cell growth via the ROS/autophagy pathway in hepatocellular carcinoma. Cancer Lett. 2011, 312, 55–61. https://doi.org/10.1016/j.canlet.2011.07.024.
- 169.
Chen, Y.; Yang, Y.; Miller, M.L.; et al. Hepatocyte-specific Gclc deletion leads to rapid onset of steatosis with mitochondrial injury and liver failure. Hepatology 2007, 45, 1118–1128.
- 170.
Orlowska, K.; Fling, R.; Nault, R.; et al. Cystine/Glutamate Xc– Antiporter Induction Compensates for Transsulfuration Pathway Repression by 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) to Ensure Cysteine for Hepatic Glutathione Biosynthesis. Chem. Res. Toxicol. 2023, 36, 900–915. https://doi.org/10.1021/acs.chemrestox.3c00017.
- 171.
Su, H.; Huang, J.; Weng, S.; et al. Glutathione synthesis primes monocytes metabolic and epigenetic pathway for β-glucan-trained immunity. Redox Biol. 2021, 48, 102206. https://doi.org/10.1016/j.redox.2021.102206.
- 172.
Lu, S. Dysregulation of glutathione synthesis in liver disease. Liver Res. 2020, 4, 64–73. https://doi.org/10.1016/j.livres.2020.05.003.
- 173.
Maddocks, O.D.K.; Berkers, C.R.; Mason, S.M.; et al. Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature 2013, 493, 542–546. https://doi.org/10.1038/nature11743.
- 174.
Yang, M.; Vousden, K.H. Serine and one-carbon metabolism in cancer. Nat. Rev. Cancer 2016, 16, 650–662. https://doi.org/10.1038/nrc.2016.81.
- 175.
Wang, K.; Luo, L.; Fu, S.; et al. PHGDH arginine methylation by PRMT1 promotes serine synthesis and represents a therapeutic vulnerability in hepatocellular carcinoma. Nat. Commun. 2023, 14, 1011. https://doi.org/10.1038/s41467-023-36708-5.
- 176.
Woo, C.C.; Chen, W.C.; Teo, X.; et al. Downregulating serine hydroxymethyltransferase 2 (SHMT2) suppresses tumorigenesis in human hepatocellular carcinoma. Oncotarget 2016, 7, 53005–53017. https://doi.org/10.18632/oncotarget.10415.
- 177.
Ji, L.; Tang, Y.; Pang, X.; et al. Increased Expression of Serine Hydroxymethyltransferase 2 (SHMT2) is a Negative Prognostic Marker in Patients with Hepatocellular Carcinoma and is Associated with Proliferation of HepG2 Cells. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2019, 25, 5823–5832. https://doi.org/10.12659/MSM.915754.
- 178.
Zhou, Q.; Li, L.; Sha, F.; et al. PTTG1 Reprograms Asparagine Metabolism to Promote Hepatocellular Carcinoma Progression. Cancer Res. 2023, 83, 2372–2386. https://doi.org/10.1158/0008-5472.CAN-22-3561.
- 179.
Zhang, B.; Dong, L.W.; Tan, Y.X.; et al. Asparagine synthetase is an independent predictor of surgical survival and a potential therapeutic target in hepatocellular carcinoma. Br. J. Cancer 2013, 109, 14–23. https://doi.org/10.1038/bjc.2013.293.
- 180.
Krall, A.S.; Xu, S.; Graeber, T.G.; et al. Asparagine promotes cancer cell proliferation through use as an amino acid exchange factor. Nat. Commun. 2016, 7, 11457. https://doi.org/10.1038/ncomms11457.
- 181.
Holeček, M. Roles of malate and aspartate in gluconeogenesis in various physiological and pathological states. Metabolism 2023, 145, 155614. https://doi.org/10.1016/j.metabol.2023.155614.
- 182.
Li, Y.; Li, B.; Xu, Y.; et al. GOT2 Silencing Promotes Reprogramming of Glutamine Metabolism and Sensitizes Hepatocellular Carcinoma to Glutaminase Inhibitors. Cancer Res. 2022, 82, 3223–3235. https://doi.org/10.1158/0008-5472.Can-22-0042.
- 183.
Cao, Y.; Ding, W.; Zhang, J.; et al. Significant Down-Regulation of Urea Cycle Generates Clinically Relevant Proteomic Signature in Hepatocellular Carcinoma Patients with Macrovascular Invasion. J. Proteome Res. 2019, 18, 2032–2044. https://doi.org/10.1021/acs.jproteome.8b00921.
- 184.
Garcia-Bermudez, J.; Baudrier, L.; La, K.; et al. Aspartate is a limiting metabolite for cancer cell proliferation under hypoxia and in tumours. Nat. Cell Biol. 2018, 20, 775–781. https://doi.org/10.1038/s41556-018-0118-z.
- 185.
Sullivan, L.B.; Luengo, A.; Danai, L.V.; et al. Aspartate is an endogenous metabolic limitation for tumour growth. Nat. Cell Biol. 2018, 20, 782–788. https://doi.org/10.1038/s41556-018-0125-0.
- 186.
Shi, J.; Wen, K.; Mui, S.; et al. Integrated analysis reveals an aspartate metabolism-related gene signature for predicting the overall survival in patients with hepatocellular carcinoma. Clin. Transl. Oncol. 2024, 26, 2181–2197. https://doi.org/10.1007/s12094-024-03431-6.
- 187.
Schlessinger, J. Cell signaling by receptor tyrosine kinases. Cell 2000, 103, 211–225. https://doi.org/10.1016/s0092-8674(00)00114-8.
- 188.
Holme, E.; Mitchell, G.A. Tyrosine Metabolism. In Physician’s Guide to the Diagnosis, Treatment, and Follow-Up of Inherited Metabolic Diseases; Blau, N., Duran, M., Gibson, K.M.; et al., Eds.; Springer: Berlin, Germany, 2014; pp. 23–31.
- 189.
Fu, L.; Dong, S.S.; Xie, Y.W.; et al. Down-regulation of tyrosine aminotransferase at a frequently deleted region 16q22 contributes to the pathogenesis of hepatocellular carcinoma. Hepatology 2010, 51, 1624–1634. https://doi.org/10.1002/hep.23540.
- 190.
Nguyen, T.N.; Nguyen, H.Q.; Le, D.-H. Unveiling prognostics biomarkers of tyrosine metabolism reprogramming in liver cancer by cross-platform gene expression analyses. PLoS ONE 2020, 15, e0229276. https://doi.org/10.1371/journal.pone.0229276.
- 191.
Yang, X.; Chen, S.-L.; Lin, C.-S.; et al. Tyrosine metabolic enzyme HPD is decreased and predicts unfavorable outcomes in hepatocellular carcinoma. Pathol. Res. Pract. 2020, 216, 153153. https://doi.org/10.1016/j.prp.2020.153153.
- 192.
Tong, M.; Wong, T.-L.; Zhao, H.; et al. Loss of tyrosine catabolic enzyme HPD promotes glutamine anaplerosis through mTOR signaling in liver cancer. Cell Rep. 2021, 36, 109617. https://doi.org/10.1016/j.celrep.2021.109617.
- 193.
Yang, F.; Li, J.; Deng, H.; et al. GSTZ1-1 Deficiency Activates NRF2/IGF1R Axis in HCC via Accumulation of Oncometabolite Succinylacetone. Embo J. 2019, 38, e101964. https://doi.org/10.15252/embj.2019101964.
- 194.
Wang, Q.; Bin, C.; Xue, Q.; et al. GSTZ1 sensitizes hepatocellular carcinoma cells to sorafenib-induced ferroptosis via inhibition of NRF2/GPX4 axis. Cell Death Dis. 2021, 12, 426. https://doi.org/10.1038/s41419-021-03718-4.
- 195.
Seibt, T.M.; Proneth, B.; Conrad, M. Role of GPX4 in ferroptosis and its pharmacological implication. Free Radic. Biol. Med. 2019, 133, 144–152. https://doi.org/10.1016/j.freeradbiomed.2018.09.014.
- 196.
Yu, X.; Long, Y.C. Crosstalk between cystine and glutathione is critical for the regulation of amino acid signaling pathways and ferroptosis. Sci. Rep. 2016, 6, 30033. https://doi.org/10.1038/srep30033.
- 197.
Gao, M.; Monian, P.; Quadri, N.; et al. Glutaminolysis and Transferrin Regulate Ferroptosis. Mol. Cell 2015, 59, 298–308. https://doi.org/10.1016/j.molcel.2015.06.011.
- 198.
Chen, X.; Kang, R.; Kroemer, G.; et al. Broadening horizons: The role of ferroptosis in cancer. Nat. Rev. Clin. Oncol. 2021, 18, 280–296.
- 199.
Zhang, H.; Wang, J.; Xiang, X.; et al. An Esterase-Responsive SLC7A11 shRNA Delivery System Induced Ferroptosis and Suppressed Hepatocellular Carcinoma Progression. Pharmaceutics 2024, 16, 249. https://doi.org/10.3390/pharmaceutics16020249.
- 200.
Wang, L.; Tong, L.; Xiong, Z.; et al. Ferroptosis-inducing nanomedicine and targeted short peptide for synergistic treatment of hepatocellular carcinoma. J. Nanobiotechnol. 2024, 22, 533. https://doi.org/10.1186/s12951-024-02808-7.
- 201.
Guo, M.; Chen, S.; Sun, J.; et al. PIP5K1A Suppresses Ferroptosis and Induces Sorafenib Resistance by Stabilizing NRF2 in Hepatocellular Carcinoma. Adv. Sci. 2025, 12, e04372. https://doi.org/10.1002/advs.202504372.
- 202.
Yan, Y.; Hu, J.; Han, N.; et al. Sorafenib-loaded metal-organic framework nanoparticles for anti-hepatocellular carcinoma effects through synergistically potentiating ferroptosis and remodeling tumor immune microenvironment. Mater. Today Bio 2025, 32, 101848. https://doi.org/10.1016/j.mtbio.2025.101848.
- 203.
Zhao, C.; Qin, G.; Ling, C.; et al. MSNs-loaded HMME and Erastin-mediated ferroptosis combined with sonodynamic therapy for HCC treatment. J. Cancer Res. Ther. 2025, 21, 465–476.
- 204.
Nie, D.; Guo, T.; Zong, X.; et al. Induction of ferroptosis by artesunate nanoparticles is an effective therapeutic strategy for hepatocellular carcinoma. Cancer Nanotechnol. 2023, 14, 81. https://doi.org/10.1186/s12645-023-00232-4.
- 205.
Liu, J.; Li, X.; Chen, J.; et al. Arsenic-Loaded Biomimetic Iron Oxide Nanoparticles for Enhanced Ferroptosis-Inducing Therapy of Hepatocellular Carcinoma. ACS Appl. Mater. Interfaces 2023, 15, 6260–6273. https://doi.org/10.1021/acsami.2c14962.
- 206.
Xuan, F.; Zhao, X.; Pang, W.; et al. Biomimetic Co-delivery of Lenvatinib and FePt Nanoparticles for Enhanced Ferroptosis/Apoptosis Treatment of Hepatocellular Carcinoma. Adv. Heal. Mater. 2025, 14, e2401747. https://doi.org/10.1002/adhm.202401747.
- 207.
Li, S.; Xu, L.; Wu, G.; et al. Remodeling Serine Synthesis and Metabolism via Nanoparticles (NPs)-Mediated CFL1 Silencing to Enhance the Sensitivity of Hepatocellular Carcinoma to Sorafenib. Adv. Sci. 2023, 10, 2207118. https://doi.org/10.1002/advs.202207118.
- 208.
Cheng, P.N.; Lam, T.L.; Lam, W.M.; et al. Pegylated recombinant human arginase (rhArg-peg5,000mw) inhibits the in vitro and in vivo proliferation of human hepatocellular carcinoma through arginine depletion. Cancer Res. 2007, 67, 309–317. https://doi.org/10.1158/0008-5472.Can-06-1945.
- 209.
Feun, L.; Savaraj, N. Pegylated arginine deiminase: A novel anticancer enzyme agent. Expert. Opin. Investig. Drugs 2006, 15, 815–822. https://doi.org/10.1517/13543784.15.7.815.
- 210.
Yau, T.; Cheng, P.N.; Chan, P.; et al. Preliminary efficacy, safety, pharmacokinetics, pharmacodynamics and quality of life study of pegylated recombinant human arginase 1 in patients with advanced hepatocellular carcinoma. Investig. New Drugs 2015, 33, 496–504. https://doi.org/10.1007/s10637-014-0200-8.
- 211.
Mousa, A.M.; Abdelraof, M.; Hassabo, A.A.; et al. Marine L-arginase encapsulated in poly (lactic-co-glycolic acid) nanoparticles: A novel anti-cancer strategy for effective hepatocellular carcinoma treatment in vitro and in vivo. J. Drug Deliv. Sci. Technol. 2025, 109, 106980. https://doi.org/10.1016/j.jddst.2025.106980.
- 212.
Fu, Y.J.; Liu, S.S.; Rodrigues, R.M.; et al. Activation of VIPR1 suppresses hepatocellular carcinoma progression by regulating arginine and pyrimidine metabolism. Int. J. Biol. Sci. 2022, 18, 4341–4356. https://doi.org/10.7150/ijbs.71134.
- 213.
Banik, D.; Noonepalle, S.; Hadley, M.; et al. HDAC6 plays a noncanonical role in the regulation of antitumor immune responses, dissemination, and invasiveness of breast cancer. Cancer Res. 2020, 80, 3649–3662.
- 214.
Wang, X.; Yan, B.; Li, H.; et al. Reprogrammed IDO-Induced Immunosuppressive Microenvironment Synergizes with Immunogenic Magnetothermodynamics for Improved Cancer Therapy. ACS Appl. Mater. Interfaces 2024, 16, 30671–30684. https://doi.org/10.1021/acsami.4c02740.
- 215.
Yang, X.; Zhang, W.; Jiang, W.; et al. Nanoconjugates to enhance PDT-mediated cancerimmunotherapy by targeting the indoleamine-2,3-dioxygenase pathway. J. Nanobiotechnol. 2021, 19, 182. https://doi.org/10.1186/s12951-021-00919-z.
- 216.
Zou, T.; Huang, Y.; Zhou, Z.; et al. A minimalist multifunctional nano-prodrug for drug resistance reverse and integration with PD-L1 mAb for enhanced immunotherapy of hepatocellular carcinoma. J. Nanobiotechnol. 2024, 22, 750. https://doi.org/10.1186/s12951-024-03027-w.
- 217.
Zhang, Y.; Feng, Y.; Huang, Y.; et al. Tumor-Targeted Gene Silencing IDO Synergizes PTT-Induced Apoptosis and Enhances Anti-tumor Immunity. Front. Immunol. 2020, 11, 968. https://doi.org/10.3389/fimmu.2020.00968.
- 218.
Wang, Y.; Zhang, B.; Xi, Q.; et al. Gemcitabine nano-prodrug reprograms intratumoral metabolism and alleviates immunosuppression for hepatocellular carcinoma therapy. Nano Today 2023, 53, 102009. https://doi.org/10.1016/j.nantod.2023.102009.
- 219.
Ghosh, A.; Ghosh, A.K.; Zaman, A.; et al. Metformin-Loaded Hyaluronic Acid-Derived Carbon Dots for Targeted Therapy against Hepatocellular Carcinoma by Glutamine Metabolic Reprogramming. Mol. Pharm. 2023, 20, 6391–6406. https://doi.org/10.1021/acs.molpharmaceut.3c00772.
- 220.
Hu, H.; Ning, S.; Liu, F.; et al. Hafnium Metal–Organic Framework-Based Glutamine Metabolism Disruptor For Potentiating Radio-Immunotherapy in MYC-Amplified Hepatocellular Carcinoma. ACS Appl. Mater. Interfaces 2025, 17, 19367–19381. https://doi.org/10.1021/acsami.4c21998.
- 221.
Brown, H.; Piknova, B.; Park, J.; et al. Comparing Nitric Oxide Generation Pathways using Dietary L-Arginine and Dietary Nitrate: A 15N tracer study in Wistar Rats. Physiology 2024, 39, 1628. https://doi.org/10.1152/physiol.2024.39.s1.1628.
- 222.
Nishikawa, M.; Sato, E.F.; Kuroki, T.; et al. Macrophage–Derived Nitric Oxide Induces Apoptosis of Rat Hepatoma CellsIn Vivo. Hepatology 1998, 28, 1474–1480.
- 223.
Hu, Y.; Xing, Y.-W.; Fan, G.; et al. L-arginine combination with 5-fluorouracil inhibit hepatocellular carcinoma cells through suppressing iNOS/NO/AKT-mediated glycolysis. Front. Pharmacol. 2024, 15, 1391636. https://doi.org/10.3389/fphar.2024.1391636.
- 224.
Yin, X.-Y.; Jiang, J.-M.; Liu, J.-Y.; et al. Effects of endogenous nitric oxide induced by 5-fluorouracil and L-Arg on liver carcinoma in nude mice. World J. Gastroenterol. 2007, 13, 6249–6253. https://doi.org/10.3748/WJG.V13.I46.6249.
- 225.
Jiang, W.; Dong, W.; Li, M.; et al. Nitric Oxide Induces Immunogenic Cell Death and Potentiates Cancer Immunotherapy. ACS Nano 2022, 16, 3881–3894. https://doi.org/10.1021/acsnano.1c09048.
- 226.
Mei, T.; Zhang, X.; Hou, X.; et al. Enhanced cancer immunotherapy via synergistic action of NO-Donor nanoparticles (NanoARG) and PD-1 antibody. Sci. Technol. Adv. Mater. 2025, 26, 2538430. https://doi.org/10.1080/14686996.2025.2538430.
- 227.
Chen, D.; Liu, X.; Lu, X.; et al. Nanoparticle drug delivery systems for synergistic delivery of tumor therapy. Front. Pharmacol. 2023, 14, 1111991. https://doi.org/10.3389/fphar.2023.1111991.
- 228.
Hou, X.; Zaks, T.; Langer, R.; et al. Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater. 2021, 6, 1078–1094. https://doi.org/10.1038/s41578-021-00358-0.
- 229.
Hosseini-Kharat, M.; Bremmell, K.E.; Prestidge, C.A. Why do lipid nanoparticles target the liver? Understanding of biodistribution and liver-specific tropism. Mol. Ther. Methods Clin. Dev. 2025, 33, 101436. https://doi.org/10.1016/j.omtm.2025.101436.
- 230.
Wang, J.; Ding, Y.; Chong, K.; et al. Recent Advances in Lipid Nanoparticles and Their Safety Concerns for mRNA Delivery. Vaccines 2024, 12, 1148. https://doi.org/10.3390/vaccines12101148.
- 231.
Stevanović, M.M.; Qian, K.; Huang, L.; et al. PLGA-Based Co-Delivery Nanoformulations: Overview, Strategies, and Recent Advances. Pharmaceutics 2025, 17, 1613. https://doi.org/10.3390/pharmaceutics17121613.
- 232.
Omidian, H.; Wilson, R.L.; Castejon, A.M. Recent Advances in Peptide-Loaded PLGA Nanocarriers for Drug Delivery and Regenerative Medicine. Pharmaceuticals 2025, 18, 127. https://doi.org/10.3390/ph18010127.
- 233.
Guo, Z.; Xiao, Y.; Wu, W.; et al. Metal-organic framework-based smart stimuli-responsive drug delivery systems for cancer therapy: Advances, challenges, and future perspectives. J. Nanobiotechnol. 2025, 23, 157. https://doi.org/10.1186/s12951-025-03252-x.

This work is licensed under a Creative Commons Attribution 4.0 International License.


