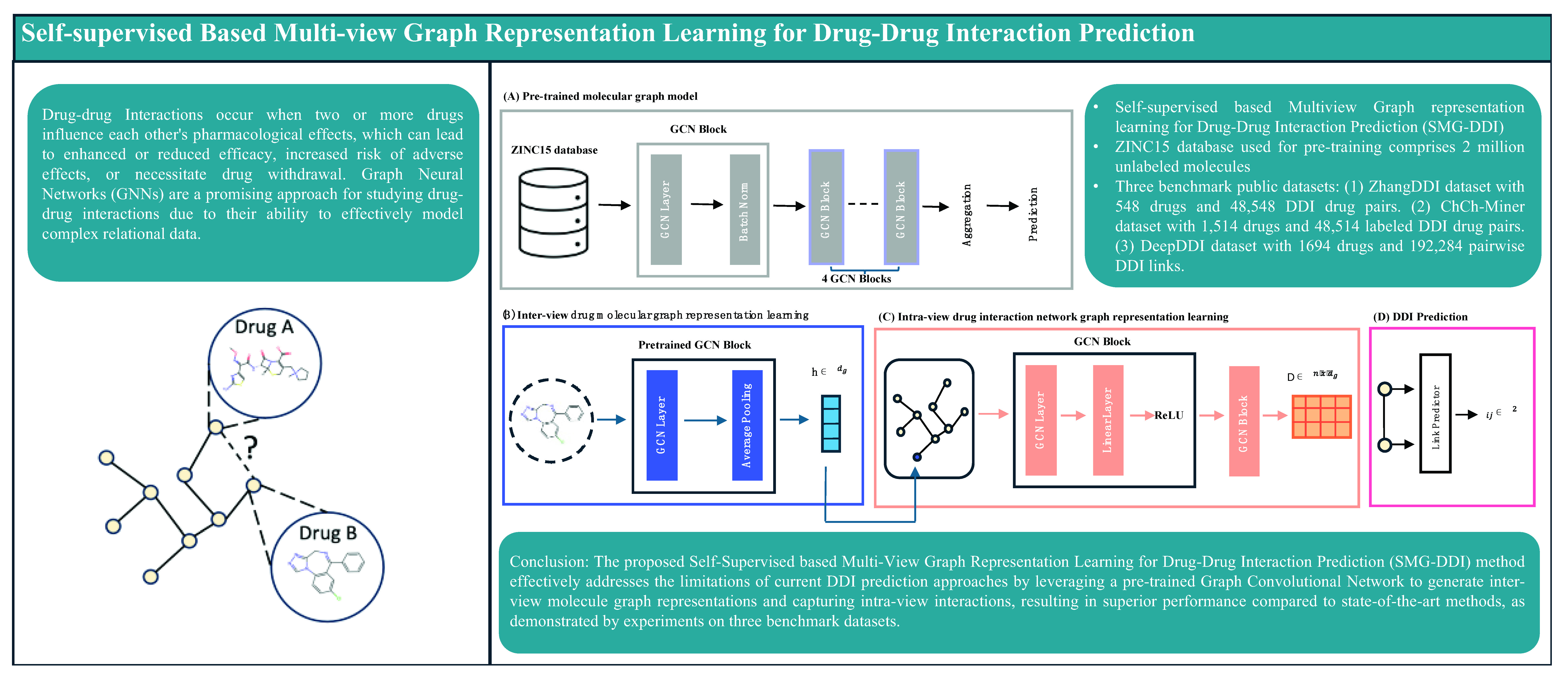
Drug-Drug Interactions (DDIs) can occur when diseases are treated with combinations of drugs, leading to changes in the pharmacological activity of these drugs. Predicting DDIs has become a crucial task in medical health. Recently, hierarchical graph representation learning methods have attracted significant interest and have proven effective for this task. However, collecting drug interaction data through biological experiments in wet laboratories is resource- and time-intensive. Given the limited amount of available drug interaction data, the performance of existing hierarchical graph methods has encountered a bottleneck. Current approaches are supervised learning methods, which train graph neural networks on specific datasets and can cause overfitting problems. Additionally, supervised learning models cannot leverage information from massive amounts of unlabeled public molecular datasets, such as ZINC15. To overcome this limitation, we propose a novel method for multi-view graph representation learning, namely, Self-Supervised Multi-View Graph Representation Learning for Drug-Drug Interaction Prediction (SMG-DDI). SMG-DDI leverages a pre-trained Graph Convolutional Network to generate inter-view molecule graph representations, incorporating atoms as nodes and chemical bonds as edges. Subsequently, SMG-DDI captures intra-view interactions between molecules. The final drug-drug interactions will be based on the drug embeddings from intra-view analyses. Our experiments conducted on various real datasets demonstrate that molecular structure information can aid in predicting potential drug-drug interactions, and our proposed approach outperforms state-of-the-art DDI prediction methods. The accuracies are 0.83, 0.79, and 0.73 on small, medium, and large scale test datasets, respectively.
- Open Access
- Article
Self-Supervised Based Multi-View Graph Presentation Learning for Drug-Drug Interaction Prediction
- Kuang Du 1,
- Jing Du 2,
- Zhi Wei 1, *
Author Information
Received: 19 Sep 2024 | Revised: 11 Oct 2024 | Accepted: 18 Oct 2024 | Published: 23 Oct 2024
Abstract
Graphical Abstract
Keywords
hierarchical graph representation learning | self-supervised learning | drug-drug interaction | molecular structural information
References
- 1.Baxter, K.; Preston, C.L. Stockley’s Drug Interactions; Pharmaceutical Press: London, UK, 2010.
- 2.Hales, C.M.; Servais, J.; Martin, C.B.; Kohen, D. Prescription Drug Use among Adults Aged 40–79 in the United States and Canada; CDC: Atlanta, GA, USA, 2019.
- 3.Kim, Y.; Zheng, S.; Tang, J.; Zheng, J.W.; Li, Z.; Jiang, X. Anticancer drug synergy prediction in understudied tissues using transfer learning. J. Am. Med. Inform. Assoc. 2021, 28, 42–51.
- 4.Vilar, S.; Uriarte, E.; Santana, L.; Lorberbaum, T.; Hripcsak, G.; Friedman, C.; Tatonetti, N.P. Similarity-based modeling in large-scale prediction of drug-drug interactions. Nat. Protoc. 2014, 9, 2147–2163.
- 5.Wen, M.; Zhang, Z.; Niu, S.; Sha, H.; Yang, R.; Yun, Y.; Lu, H. Deep-Learning-Based Drug–Target Interaction Prediction. J. Proteome Res. 2017, 16, 1401–1409. https://doi.org/10.1021/acs.jproteome.6b00618.
- 6.You, J.; McLeod, R.D.; Hu, P. Predicting drug-target interaction network using deep learning model. Comput. Biol. Chem. 2019, 80, 90–101. https://doi.org/10.1016/j.compbiolchem.2019.03.016.
- 7.Ryu, J.Y.; Kim, H.U.; Lee, S.Y. Deep learning improves prediction of drug–drug and drug–food interactions. Proc. Natl. Acad. Sci. USA 2018, 115, E4304–E4311.
- 8.Gilmer, J.; Schoenholz, S.S.; Riley, P.F.; Vinyals, O.; Dahl, G.E. Neural message passing for quantum chemistry. In Proceedings of the International Conference on Machine Learning, Sydney, Australi, 6 August 2017.
- 9.Feng, Y.-H.; Zhang, S.-W. Prediction of drug-drug interaction using an attention-based graph neural network on drug molecular graphs. Molecules 2022, 27, 3004.
- 10.Bai, Y.; Gu, K.; Sun, Y.; Wang, W. Bi-level graph neural networks for drug-drug interaction prediction. arXiv 2020, arXiv:2006.14002.
- 11.Wang, Y.; Min, Y.; Chen, X.; Wu, J. Multi-view graph contrastive representation learning for drug-drug interaction prediction. Proc. Web Conf. 2021, 2021, 2921–2933.
- 12.Wang, H.; Kaddour, J.; Liu, S.; Tang, J.; Lasenby, J.; Liu, Q. Evaluating self-supervised learning for molecular graph embeddings. arXiv 2022, arXiv:2206.08005.
- 13.You, Y.; Chen, T.; Wang, Z.; Shen, Y. When does self-supervision help graph convolutional networks? In Proceedings of the International Conference on Machine Learning, Online, 21 November 2020.
- 14.Wu, L.; Lin, H.; Tan, C.; Gao, Z.; Li, S.Z. Self-supervised learning on graphs: Contrastive, generative, or predictive. IEEE Trans. Knowl. Data Eng. 2021, 35, 4216–4235.
- 15.Liu, S.; Wang, H.; Liu, W.; Lasenby, J.; Guo, H.; Tang, J. Pre-training molecular graph representation with 3d geometry. arXiv 2021, arXiv:2110.07728.
- 16.Hu, W.; Liu, B.; Gomes, J.; Zitnik, M.; Liang, P.; Pande, V.; Leskovec, J. Strategies for pre-training graph neural networks. arXiv 2019, arXiv:1905.12265.
- 17.Wu, Z.; Ramsundar, B.; Feinberg, E.N.; Gomes, J.; Geniesse, C.; Pappu, A.S.; Leswing, K.; Pande, V. MoleculeNet: A benchmark for molecular machine learning. Chem. Sci. 2018, 9, 513–530.
- 18.Zitnik, M.; Sosič, R.; Feldman, M.W.; Leskovec, J. Evolution of resilience in protein interactomes across the tree of life. Proc. Natl. Acad. Sci. USA 2019, 116, 4426–4433.
- 19.Ramsundar, B.; Eastman, P.; Walters, P.; Pande, V. Deep Learning for the Life Sciences: Applying Deep Learning to Genomics, Microscopy, Drug Discovery, and More; O’Reilly Media, Inc.: Sebastopol, CA, USA, 2019.
- 20.Bemis, G.W.; Murcko, M.A. The properties of known drugs. 1. Molecular frameworks. J. Med. Chem. 1996, 39, 2887–2893.
- 21.Chen, B.; Sheridan, R.P.; Hornak, V.; Voigt, J.H. Comparison of Random Forest and Pipeline Pilot Naïve Bayes in Prospective QSAR Predictions. J. Chem. Inf. Model. 2012, 52, 792–803. https://doi.org/10.1021/ci200615h.
- 22.Sheridan, R.P. Time-Split Cross-Validation as a Method for Estimating the Goodness of Prospective Prediction. J. Chem. Inf. Model. 2013, 53, 783–790. https://doi.org/10.1021/ci400084k.
- 23.Zellinger, W.; Grubinger, T.; Lughofer, E.; Natschläger, T.; Saminger-Platz, S. Central Moment Discrepancy (CMD) for Domain-Invariant Representation Learning. arXiv 2017, arXiv:1702.08811.
- 24.Landrum, G. Rdkit: A software suite for cheminformatics, computational chemistry, and predictive modeling. Greg Landrum 2013, 8, 5281.
- 25.Xu, Y.; Ma, J.; Liaw, A.; Sheridan, R.P.; Svetnik, V. Demystifying Multitask Deep Neural Networks for Quantitative Structure–Activity Relationships. J. Chem. Inf. Model. 2017, 57, 2490–2504. https://doi.org/10.1021/acs.jcim.7b00087.
- 26.Ching, T.; Himmelstein, D.S.; Beaulieu-Jones, B.K.; Kalinin, A.A.; Brian, T.D.; Gregory, P.W.; Ferrero, E.; Agapow, P.-M.; Zietz, M.; Hoffman, M.M.; et al. Opportunities and Obstacles for Deep Learning in Biology and Medicine. J. R. Soc. Interface 2018, 15, 20170387. https://doi.org/10.1098/rsif.2017.0387.
- 27.Wang, J.; Agarwal, D.; Huang, M.; Hu, G.; Zhou, Z.; Ye, C.; Zhang, N.R. Data denoising with transfer learning in single-cell transcriptomics. Nat. Methods 2019, 16, 875–878. https://doi.org/10.1038/s41592-019-0537-1.
- 28.Sterling, T.; Irwin, J.J. ZINC 15--Ligand Discovery for Everyone. J. Chem. Inf. Model. 2015, 55, 2324–2337. https://doi.org/10.1021/acs.jcim.5b00559.
- 29.Mikolov, T.; Sutskever, I.; Chen, K.; Corrado, G.S.; Dean, J. Distributed Representations of Words and Phrases and Their Compositionality; Curran Associates, Inc.: San Francisco, CA, USA, 2013.
- 30.Hamilton, W.; Ying, Z.; Leskovec, J. Inductive Representation Learning on Large Graphs; Curran Associates, Inc.: San Francisco, CA, USA, 2017.
- 31.Devlin, J.; Chang, M.-W.; Lee, K.; Toutanova, K. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. arXiv 2018, arXiv:1810.04805.
- 32.Zhang, W.; Chen, Y.; Liu, F.; Luo, F.; Tian, G.; Li, X. Predicting potential drug-drug interactions by integrating chemical, biological, phenotypic and network data. BMC Bioinform. 2017, 18, 18. https://doi.org/10.1186/s12859-016-1415-9.
- 33.Zitnik, R.S.M.; Maheshwari, S.; Leskovec, J. BioSNAP Datasets: Stanford Biomedical Network Dataset Collection. Available online: http://snap.stanford.edu/biodata (accessed on 1 August 2018).
- 34.Weininger, D. SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J. Chem. Inf. Comput. Sci. 1988, 28, 31–36.
- 35.Kipf, T.N.; Welling, M. Semi-Supervised Classification with Graph Convolutional Networks. arXiv 2016, arXiv:1609.02907.
- 36.Xu, K.; Hu, W.; Leskovec, J.; Jegelka, S. How Powerful are Graph Neural Networks? arXiv 2018, arXiv:1810.00826.
- 37.Hamilton, W.; Ying, Z.; Leskovec, J. Inductive Representation Learning on Large Graphs; Curran Associates, Inc.: San Francisco, CA, USA, 2017.
- 38.Li, J.; Rong, Y.; Cheng, H.; Meng, H.; Huang, W.; Huang, J. Semi-Supervised Graph Classification: A Hierarchical Graph Perspective. In Proceedings of the World Wide Web Conference, San Francisco, CA, USA, 13–17 May 2019.
- 39.Ross, J.; Belgodere, B.; Chenthamarakshan, V.; Padhi, I.; Mroueh, Y.; Das, P. Large-scale chemical language representations capture molecular structure and properties. Nat. Mach. Intell. 2022, 4, 1256–1264. https://doi.org/10.1038/s42256-022-00580-7.
How to Cite
Du, K.; Du, J.; Wei, Z. Self-Supervised Based Multi-View Graph Presentation Learning for Drug-Drug Interaction Prediction. Transactions on Artificial Intelligence 2025, 1 (1), 1–15. https://doi.org/10.53941/tai.2025.100001.
RIS
BibTex
Copyright & License

Transactions on Artificial Intelligence
This work is licensed under a Creative Commons Attribution 4.0 International License.
Contents
References


