Drug discovery and development is a complex and expensive process. There are two approaches, phenotypic and target-based approaches, each holding different advantages for screening novel drug candidates when pursuing successful marketing authorization. However, the attrition rates of drug candidates continue to increase. In this review, we discuss recent successes and ongoing advances in phenotypic screening and target-based screening for drug discovery. We also explore how strategic and technological innovations may fuel new approaches in drug discovery. There are two approaches in drug discovery.
- Open Access
- Review
Advances of the Target-Based and Phenotypic Screenings and Strategies in Drug Discovery
- Shoubao Wang 1, *,
- Zihan Wang 1,
- Lianhua Fang 1,
- Yang Lv 2,
- Guanhua Du 1, *
Author Information
Received: 08 Nov 2022 | Accepted: 10 Dec 2022 | Published: 21 Dec 2022
Abstract
Graphical Abstract
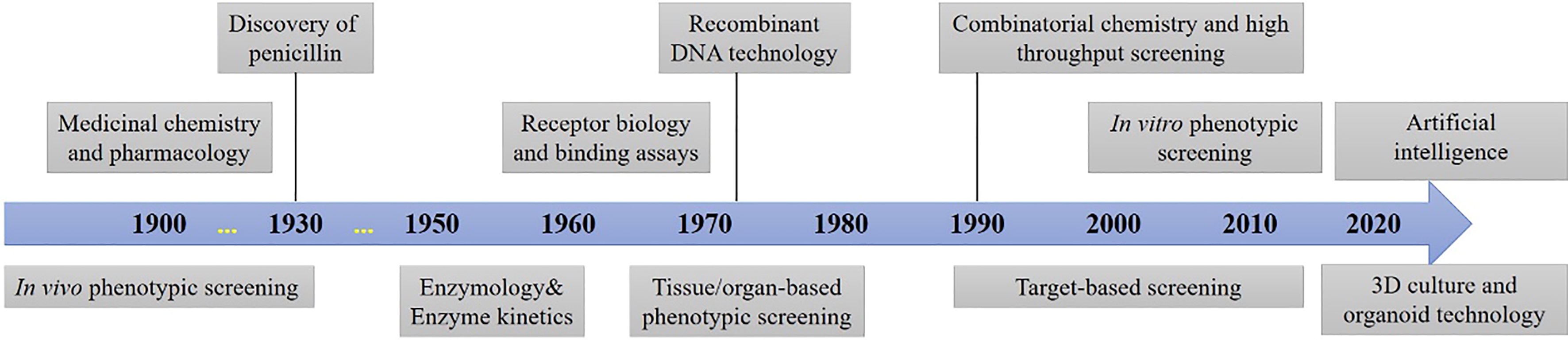
Keywords
drug discovery | phenotypic approach | target-based approach | screening
References
- 1.Eder J.; Herrling P.L. Trends in modern drug discovery. Handb. Exp. Pharmacol., 2016, 232: 3-22.
- 2.Moffat J.G.; Vincent F.; Lee J.A.; et al. Opportunities and challenges in phenotypic drug discovery: an industry perspective. Nat. Rev. Drug Discovery, 2017, 16(8): 531-543.
- 3.Sams-Dodd F. Target-based drug discovery: is something wrong? Drug Discov. Today. 2005, 10(2): 139-147.
- 4.Hurko O. Target-based drug discovery, genetic diseases, and biologics. Neurochem. Int., 2012, 61(6): 892-898.
- 5.Zheng W.; Thorne N.; McKew J.C. Phenotypic screens as a renewed approach for drug discovery. Drug Discovery Today, 2013, 18(21/22): 1067-1073.
- 6.Blay V.; Tolani B.; Ho S.P.; et al. High-throughput screening: today's biochemical and cell-based approaches. Drug Discov. Today., 2020, 25(10): 1807-1821.
- 7.Stoddart L.A.; White C.W.; Nguyen K.; et al. Fluorescence- and bioluminescence-based approaches to study GPCR ligand binding. Br. J. Pharmacol., 2016, 173(20): 3028-3037.
- 8.Togre N.S; Vargas A.M.; Bhargavi G.;et al. Fragment-based drug discovery against mycobacteria: the success and challenges. Int. J. Mol. Sci., 2022, 23(18): 10669.
- 9.Parasrampuria D.A.; Benet L.Z.; Sharma A. Why drugs fail in late stages of development: case study analyses from the last decade and recommendations. AAPS J., 2018, 20(3): 46.
- 10.Yasgar A.; Jadhav A.; Simeonov A.; et al. AlphaScreen-based assays: ultra-high-throughput screening for small-molecule inhibitors of challenging enzymes and protein-protein interactions. Methods Mol. Biol., 2016, 1439: 77-98.
- 11.Zhao G.X.; Huang Y.R.; Zhou Y.; et al. Future challenges with DNA-encoded chemical libraries in the drug discovery domain. Expert Opin. Drug Discovery, 2019, 14(8): 735-753.
- 12.Ramsay R.R.; Popovic-Nikolic M.R.; Nikolic K.; et al. A perspective on multi-target drug discovery and design for complex diseases. Clin. Transl. Med., 2018, 7(1): 3.
- 13.Reddy A.S.; Zhang S.X. Polypharmacology: drug discovery for the future. Expert Rev. Clin. Pharmacol., 2013, 6(1): 41-47.
- 14.Nováček V.; Mohamed S.K. Predicting polypharmacy side-effects using knowledge graph embeddings. AMIA Joint Summits on Translational Science proceedings. AMIA Jt. Summits Transl. Sci. Proc., 2020, 2020: 449-458.
- 15.Hughes R.E.; Elliott R.J.R.; Dawson J.C.; et al. High-content phenotypic and pathway profiling to advance drug discovery in diseases of unmet need. Cell Chem. Biol., 2021, 28(3): 338-355.
- 16.Mahner M.; Kary M. What exactly are genomes, genotypes and phenotypes? And what about phenomes?. J. Theor. Biol., 1997, 186(1): 55-63.
- 17.Swinney D.C.; Anthony J. How were new medicines discovered? Nat. Rev. Drug Discovery, 2011, 10(7): 507-519.
- 18.Swinney D.C.; Lee J.A. Recent advances in phenotypic drug discovery [version 1; peer review: 2 approved]. F1000Research, 2020, 9(Faculty Rev): 944.
- 19.Szabo M.; Svensson Akusjärvi S.; Saxena A.; et al. Cell and small animal models for phenotypic drug discovery. Drug Des., Dev. Ther., 2017, 11: 1957-1967.
- 20.Vincent F.; Nueda A.; Lee J.; et al. Phenotypic drug discovery: recent successes, lessons learned and new directions. Nat. Rev. Drug Discovery, 2022, 21(12): 899-914.
- 21.Liu C.; Oikonomopoulos A.; Sayed N.; et al. Modeling human diseases with induced pluripotent stem cells: from 2D to 3D and beyond. Development, 2018, 145(5): dev156166.
- 22.Sayed N.; Liu C.; Wu J.C. Translation of human-induced pluripotent stem cells: from clinical trial in a dish to precision medicine. J. Am. Coll. Cardiol., 2016, 67(18): 2161-2176.
- 23.Corrò C.; Novellasdemunt L.; Li V.S.W. A brief history of organoids. Am. J. Physiol.: Cell Physiol., 2020, 319(1): C151-C165.
- 24.Takahashi T. Organoids for drug discovery and personalized medicine. Annu. Rev. Pharmacol. Toxicol., 2019, 59: 447-462.
- 25.Zhao Y.; Kankala R.K.; Wang S.B.; et al. Multi-organs-on-chips: towards long-term biomedical investigations. Molecules, 2019, 24(4): 675.
- 26.Ronaldson-Bouchard K.; Teles D.; Yeager K.; et al. A multi-organ chip with matured tissue niches linked by vascular flow. Nat. Biomed. Eng., 2022, 6(4): 351-371.
- 27.Paul D.; Sanap G.; Shenoy S.; et al. Artificial intelligence in drug discovery and development. Drug Discovery Today, 2021, 26(1): 80-93.
- 28.Jiménez-Luna J.; Grisoni F.; Weskamp N.; et al. Artificial intelligence in drug discovery: recent advances and future perspectives. Expert Opin. Drug Discovery, 2021, 16(9): 949-959.
- 29.Friese A.; Ursu A.; Hochheimer A.; et al. The convergence of stem cell technologies and phenotypic drug discovery. Cell Chem. Biol., 2019, 26(8): 1050-1066.
- 30.Iwata H.; Kojima R.; Okuno Y. An in silico approach for integrating phenotypic and target-based approaches in drug discovery. Mol. Inf., 2020, 39(1/2): e1900096.
- 31.Aulner N.; Danckaert A.; Ihm J.; et al. Next-generation phenotypic screening in early drug discovery for infectious diseases. Trends Parasitol., 2019, 35(7): 559-570.
How to Cite
Wang, S.; Wang, Z.; Fang, L.; Lv, Y.; Du, G. Advances of the Target-Based and Phenotypic Screenings and Strategies in Drug Discovery. International Journal of Drug Discovery and Pharmacology 2022, 1 (1), 2. https://doi.org/10.53941/ijddp.v1i1.199.
RIS
BibTex
Copyright & License

Shou-bao Wang, Zihan Wang, Lianhua Fang, Yang Lv , Guanhua Du
This work is licensed under a Creative Commons Attribution 4.0 International License.
Contents
References


