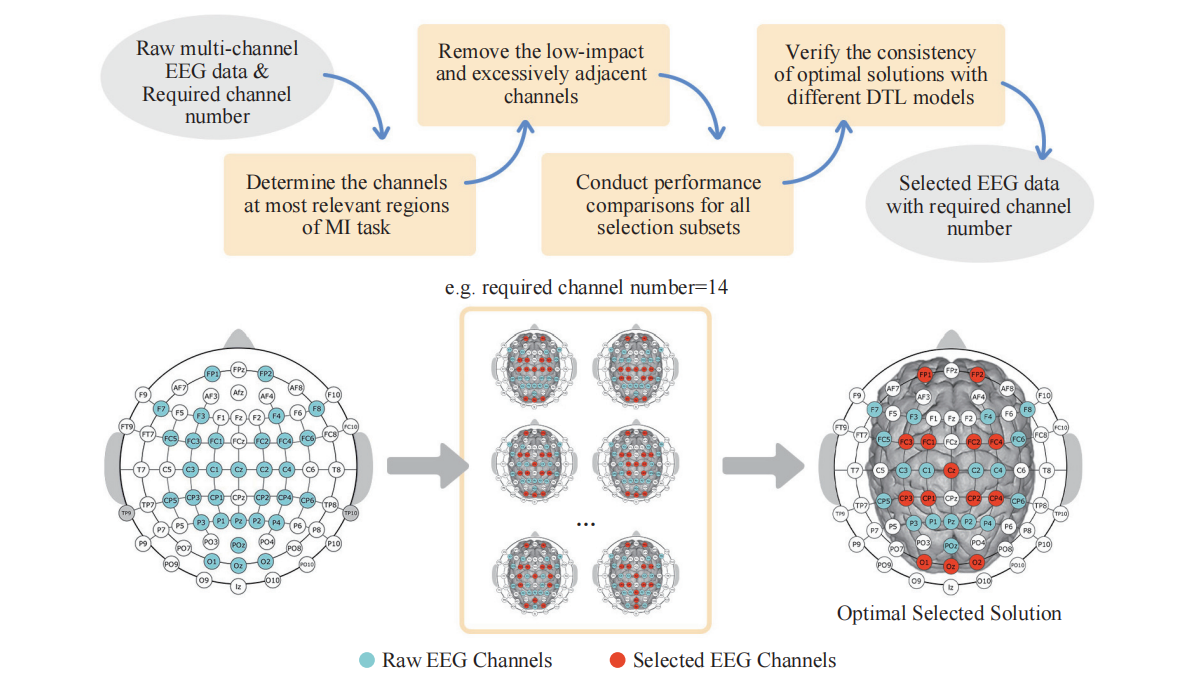
Lower limb motor imagery (MI) classification is a challenging research topic in the area of brain-computer interfaces (BCIs), and entails numerous signal channels to provide sufficient information about the background neural activity. However, practical applications often lack the environment to accommodate excessive channels due to the time-consuming setup process, inconvenient movement, and restricted application scenarios. The existing channel selection algorithms (designed for the individual subject) place a great deal of focus on the classified performance comparisons, whereas the significance of actual locations and neural functions of brain regions is disregarded. Although these algorithms require significant computation resources, their selected solutions cannot be re-used for other subjects to realize the cross-subject channel selection and improve the reusability of model due to poor interpretability and inapplicability. To date, there have been no investigations about the cross-subject channel selection problem for the lower limb MI stepping tasks. This study proposes an optimal cross-subject lower limb channel selection that selectively retains significant channels, narrows the computation scope of the selection, and obtains the optimal selection solutions. Through stepping-based MI experiments, the proposed optimal channel selection enables effective recognition in low-channel settings, thereby contributing a lot to the development of generic and convenient lower limb BCI systems. Additionally, statistical analysis reveals a significant difference in energy spectrum between left and right stepping-based MI tasks in the ![]() and
and  bands of the frontal lobe channels, providing new evidence that the frontal lobe dramatically affects lower limb MI tasks.
bands of the frontal lobe channels, providing new evidence that the frontal lobe dramatically affects lower limb MI tasks.
- Open Access
- Article
Cross-Subject EEG Channel Selection Method for Lower Limb Brain-Computer Interface
- Mingnan Wei 1, 2,
- Mengjie Huang 3, *,
- Jiaying Ni 3
Author Information
Received: 27 Apr 2023 | Accepted: 30 Jun 2023 | Published: 26 Sep 2023
Abstract
Graphical Abstract
Keywords
brain-computer interface | motor imagery | channel selection | cross-subject | deep transfer learning
References
- 1.Olsen, S.; Zhang, J.W.; Liang, K.F.; et al. An artificial intelligence that increases simulated brain–computer interface performance. J. Neural Eng., 2021, 18: 046053.
- 2.Singh, A.; Hussain, A.A.; Lal, S.; et al. A comprehensive review on critical issues and possible solutions of motor imagery based electroencephalography brain-computer interface. Sensors, 2021, 21: 2173.
- 3.Zabcikova, M.; Koudelkova, Z.; Jasek, R.; et al. Recent advances and current trends in brain-computer interface research and their applications. Int. J. Dev. Neurosci., 2022, 82: 107−123.
- 4.Ahn, M.; Cho, H.; Ahn, S.; et al. User’s self-prediction of performance in motor imagery brain-computer interface. Front. Hum. Neurosci., 2018, 12: 59.
- 5.Leeuwis, N.; Yoon, S.; Alimardani, M. Functional connectivity analysis in motor-imagery brain computer interfaces. Front. Human Neurosci., 2021, 15: 732946.
- 6.Wan, Z.T.; Yang, R.; Huang, M.; et al. EEG fading data classification based on improved manifold learning with adaptive neighborhood selection. Neurocomputing, 2022, 482: 186−196.
- 7.Wang, H.T.; Li, T.; Bezerianos, A.; et al. The control of a virtual automatic car based on multiple patterns of motor imagery BCI. Med. Biol. Eng. Comput., 2019, 57: 299−309.
- 8.Yu, N.X.; Yang, R.; Huang, M.J. Deep common spatial pattern based motor imagery classification with improved objective function. Int. J. Network Dyn. Intell., 2022, 1: 73−84.
- 9.Formaggio, E.; Masiero, S.; Bosco, A.; et al. Quantitative EEG evaluation during robot-assisted foot movement. IEEE Trans. Neural Syst. Rehabilit. Eng., 2016, 25: 1633−1640.
- 10.Kline, A.; Ghiroaga, C.G.; Pittman, D.; et al. EEG differentiates left and right imagined lower limb movement. Gait Post., 2021, 84: 148−154.
- 11.Tariq, M.; Trivailo, P.M.; Simic, M. EEG-based BCI control schemes for lower-limb assistive-robots. Front. Human Neurosci., 2018, 12: 312.
- 12.Dai, Y.X.; Wang, X.; Zhang, P.B.; et al. Sparsity constrained differential evolution enabled feature-channel-sample hybrid selection for daily-life EEG emotion recognition. Multimed. Tools Appl., 2018, 77: 21967−21994.
- 13.Geng, H.; Wang, Z.D.; Chen, Y.; et al. Multi-sensor filtering fusion with parametric uncertainties and measurement censoring: Monotonicity and boundedness. IEEE Trans. Signal Process., 2021, 69: 5875−5890.
- 14.Li, Q.; Wang, Z.D.; Shen, B.; et al. A resilient approach to recursive distributed filtering for multirate systems over sensor networks with time-correlated fading channels. IEEE Trans. Signal Inf. Process. Over Networks, 2021, 7: 636−647.
- 15.Nakanishi, M.; Wang, Y.T.; Wei, C.S.; et al. Facilitating calibration in high-speed BCI spellers via leveraging cross-device shared latent responses. IEEE Trans. Biomed. Eng., 2020, 67: 1105−1113.
- 16.Strypsteen, T.; Bertrand, A. End-to-end learnable EEG channel selection for deep neural networks with gumbel-softmax. J. Neural Eng., 2021, 18: 0460a9.
- 17.Qi, F.F.; Wu, W.; Yu, Z.L.; et al. Spatiotemporal-filtering-based channel selection for single-trial EEG classification. IEEE Trans. Cybernet., 2021, 51: 558−567.
- 18.Gurve, D.; Delisle-Rodriguez, D.; Romero-Laiseca, M.; et al. Subject-specific EEG channel selection using non-negative matrix factorization for lower-limb motor imagery recognition. J. Neural Eng., 2020, 17: 026029.
- 19.Gaur, P.; McCreadie, K.; Pachori, R.B.; et al. An automatic subject specific channel selection method for enhancing motor imagery classification in EEG-BCI using correlation. Biomed. Signal Process. Control, 2021, 68: 102574.
- 20.Homan, R.W.; Herman, J.; Purdy, P. Cerebral location of international 10–20 system electrode placement. Electroencephalogr. Clin. Neurophysiol., 1987, 66: 376−382.
- 21.Hsu, W.C.; Lin, L.F.; Chou, C.W.; et al. EEG classification of imaginary lower limb stepping movements based on fuzzy support vector machine with kernel-induced membership function. Int. J. Fuzzy Syst., 2017, 19: 566−579.
- 22.Liu, Y.H.; Lin, L.F.; Chou, C.W.; et al. Analysis of electroencephalography event-related desynchronisation and synchronisation induced by lower-limb stepping motor imagery. J. Med. Biol. Eng., 2019, 39: 54−69.
- 23.Brissenden, J.A.; Tobyne, S.M.; Osher, D.E.; et al. Topographic cortico-cerebellar networks revealed by visual attention and working memory. Curr. Biol., 2018, 28: 3364−3372.e5.
- 24.Leisman, G.; Moustafa, A.A.; Shafir, T. Thinking, walking, talking: Integratory motor and cognitive brain function. Front. Public Health, 2016, 4: 94.
- 25.Raffin, E.; Mattout, J.; Reilly, K.T.; et al. Disentangling motor execution from motor imagery with the phantom limb. Brain, 2012, 135: 582−595.
- 26.Ren, S.X.; Wang, W.Q.; Hou, Z.G.; et al. Enhanced motor imagery based brain-computer interface via FES and VR for lower limbs. IEEE Trans. Neural Syst. Rehabilit. Eng., 2020, 28: 1846−1855.
- 27.Brunner, C.; Delorme, A.; Makeig, S. EEGLAB–an open source matlab toolbox for electrophysiological research. Biomed. Tech. 2013, 58 Suppl 1, SI-1-Track-G, 000010151520134182. doi: 10.1515/bmt-2013-4182
- 28.Geng, H.; Liu, H.J.; Ma, L.F.; et al. Multi-sensor filtering fusion meets censored measurements under a constrained network environment: Advances, challenges and prospects. Int. J. Syst. Sci., 2021, 52: 3410−3436.
- 29.Mao, J.Y.; Sun, Y.; Yi, X.J.; et al. Recursive filtering of networked nonlinear systems: A survey. Int. J. Syst. Sci., 2021, 52: 1110−1128.
- 30.Debener, S.; Thorne, J.; Schneider, T.R.; et al. Using ICA for the analysis of multi-channel EEG data. In Simultaneous EEG and fMRI: Recording, Analysis, and Application; Ullsperger, M.; Debener, S., Eds.; Oxford University Press: Oxford, 2010; pp. 121–134.
- 31.He, H.; Wu, D.R. Transfer learning for brain–computer interfaces: A Euclidean space data alignment approach. IEEE Trans. Biomed. Eng., 2020, 67: 399−410.
- 32.Yang, B.H.; He, M.Y.; Liu, Y.Y.; et al. Multi-class feature extraction based on common spatial patterns of multi-band cross filter in BCIs. In International Computer Science Conference, Shanghai, China, 27–30 October 2012; Springer: Berlin/Heidelberg, Germany, 2012; pp. 399–408. doi: 10.1007/978-3-642-34381-0_46
- 33.Tan, C.Q.; Sun, F.C.; Kong, T.; et al. A survey on deep transfer learning. In 27th International Conference on Artificial Neural Networks, Rhodes, Greece, 4–7 October, 2018; Springer: Berlin/Heidelberg, Germany, 2018; pp. 270–279. doi: 10.1007/978-3-030-01424-7_27
- 34.Tan, C.Q.; Sun, F.C.; Zhang, W.C. Deep transfer learning for EEG-based brain computer interface. In 2018 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Calgary, AB, Canada, 15–20 April 2018; IEEE: New York, 2018; pp. 916–920. doi: 10.1109/ICASSP.2018.8462115
- 35.Völker, M.; Schirrmeister, R.T.; Fiederer, L.D.J.; et al. Deep transfer learning for error decoding from non-invasive EEG. In 2018 6th International Conference on Brain-Computer Interface (BCI), Gangwon, Korea (South), 15–17 January 2018; IEEE: New York, 2018; pp. 1–6. doi: 10.1109/IWW-BCI.2018.8311491
- 36.Wan, Z.T.; Yang, R.; Huang, M.J.; et al. A review on transfer learning in EEG signal analysis. Neurocomputing, 2021, 421: 1−14.
- 37.Zhao, G.Y.; Li, Y.T.; Xu, Q.R. From emotion AI to cognitive AI. Int. J. Network Dyn. Intell., 2022, 1: 65−72.
- 38.Deepak, S.; Ameer, P.M. Brain tumor classification using deep CNN features via transfer learning. Comput. Biol. Med., 2019, 111: 103345.
- 39.Pahar, M.; Klopper, M.; Warren, R.; et al. COVID-19 detection in cough, breath and speech using deep transfer learning and bottleneck features. Comput. Biol. Med., 2022, 141: 105153.
- 40.Wang, C.; Wang, Z.D.; Liu, W.B.; et al. A novel deep offline-to-online transfer learning framework for pipeline leakage detection with small samples. IEEE Trans. Instrum. Meas., 2022, 72: 3503913.
- 41.Dehghani, M.; Mobaien, A.; Boostani, R. A deep neural network-based transfer learning to enhance the performance and learning speed of BCI systems. Brain-Comput. Interfaces, 2021, 8: 14−25.
- 42.Lu, B.; Song, B.Y.; Xu, L. Human face recognition based on convolutional neural network and augmented dataset. Syst. Sci. Control Eng., 2021, 9: 29−37.
- 43.Wang, X.Y.; Yang, R.; Huang, M.J. An unsupervised deep-transfer-learning-based motor imagery EEG classification scheme for brain–computer interface. Sensors, 2022, 22: 2241.
- 44.Zeng, N.Y.; Wang, Z.D.; Zhang, H.; et al. An improved particle filter with a novel hybrid proposal distribution for quantitative analysis of gold immunochromatographic strips. IEEE Trans. Nanotechnol., 2019, 18: 819−829.
- 45.Zhang, B.C.; Wang, W.N.; Xiao, Y.T.; et al. Cross-subject seizure detection in EEGs using deep transfer learning. Comput. Math. Methods Med., 2020, 2020: 7902072.
- 46.Bagherzadeh, S.; Shahabi, M.S.; Shalbaf, A. Detection of schizophrenia using hybrid of deep learning and brain effective connectivity image from electroencephalogram signal. Comput. Biol. Med., 2022, 146: 105570.
- 47.Ding, Y.L.; Fu, M.H.; Luo, P.; et al. Network learning for biomarker discovery. Int. J. Network Dyn. Intell., 2023, 2: 51−65.
- 48.Khademi, Z.; Ebrahimi, F.; Kordy, H.M. A transfer learning-based CNN and LSTM hybrid deep learning model to classify motor imagery EEG signals. Comput. Biol. Med., 2022, 143: 105288.
- 49.Li, X.; Li, M.L.; Yan, P.F.; et al. Deep learning attention mechanism in medical image analysis: Basics and beyonds. Int. J. Network Dyn. Intell., 2023, 2: 93−116.
- 50.Shakiba, F.M.; Shojaee, M.; Azizi, S.M.; et al. Real-time sensing and fault diagnosis for transmission lines. Int. J. Network Dyn. Intell., 2022, 1: 36−47.
- 51.Alicja, M.; Maciej, S. Can AI see bias in X-ray images. Int. J. Network Dyn. Intell., 2022, 1: 48−64.
- 52.Wei, M.N.; Yang, R.; Huang, M.J. Motor imagery EEG signal classification based on deep transfer learning. In 2021 IEEE 34th International Symposium on Computer-Based Medical Systems (CBMS), Aveiro, Portugal, 7–9 June 2021; IEEE: New York, 2021; pp 85–90. doi: 10.1109/CBMS52027.2021.00083
- 53.Cheng, H.J.; Wang, Z.D.; Wei, Z.H.; et al. On adaptive learning framework for deep weighted sparse autoencoder: A multiobjective evolutionary algorithm. IEEE Trans. Cybern., 2020, 52: 3221−3231.
- 54.Liang, C.M.; Li, Y.W.; Liu, Y.H.; et al. Segmentation and weight prediction of grape ear based on SFNet-ResNet18. Syst. Sci. Control Eng., 2022, 10: 722−732.
- 55.Morid, M.A.; Borjali, A.; Del Fiol, G. A scoping review of transfer learning research on medical image analysis using ImageNet. Comput. Biol. Med., 2021, 128: 104115.
- 56.Sun, J.Y.; Wang, Z.D.; Yu, H.; et al. Two-stage deep regression enhanced depth estimation from a single RGB image. IEEE Trans. Emerg. Top. Comput., 2022, 10: 719−727.
How to Cite
Wei, M.; Huang, M.; Ni, J. Cross-Subject EEG Channel Selection Method for Lower Limb Brain-Computer Interface. International Journal of Network Dynamics and Intelligence 2023, 2 (3), 100008. https://doi.org/10.53941/ijndi.2023.100008.
RIS
BibTex
Copyright & License

Copyright (c) 2023 by the authors.
This work is licensed under a Creative Commons Attribution 4.0 International License.
Contents
References


