Metabolic dysfunction-associated steatotic liver disease (MASLD) is becoming the leading cause of chronic liver disease-related morbidity and mortality globally. It encompasses a spectrum from metabolic dysfunction-associated steatotic liver (MASL) to metabolic dysfunction-associated steatohepatitis (MASH), potentially advancing to cirrhosis and hepatocellular carcinoma. While the pathogenesis of MASLD is complicated and not yet completely elucidated, hepatic macrophages, including liver resident Kupffer cells (Res-KCs) and recruited circulating monocyte-derived macrophages (MoDMs), play a pivotal role in its initiation and progression. Recent advancements in single-cell RNA sequencing have unveiled significant heterogeneity among macrophages and their diverse contributions to MASLD progression. This review delineates the origin and surface markers of hepatic macrophages, emphasizing their multifaceted roles in the pathogenesis of MASLD in steatosis, inflammation and fibrosis. Furthermore, we delve into the latest advancements in pharmacological treatment strategies for patients with MASLD.
- Open Access
- Review
The Multifaced Roles of Hepatic Macrophages in MASLD
- Xiaoping Sui 1,
- Minqi Wu 1,
- Xue Yang 1,
- Yanan Wang 1, 2, *
Author Information
Received: 20 Sep 2024 | Revised: 14 Oct 2024 | Accepted: 24 Oct 2024 | Published: 29 Oct 2024
Abstract
Graphical Abstract
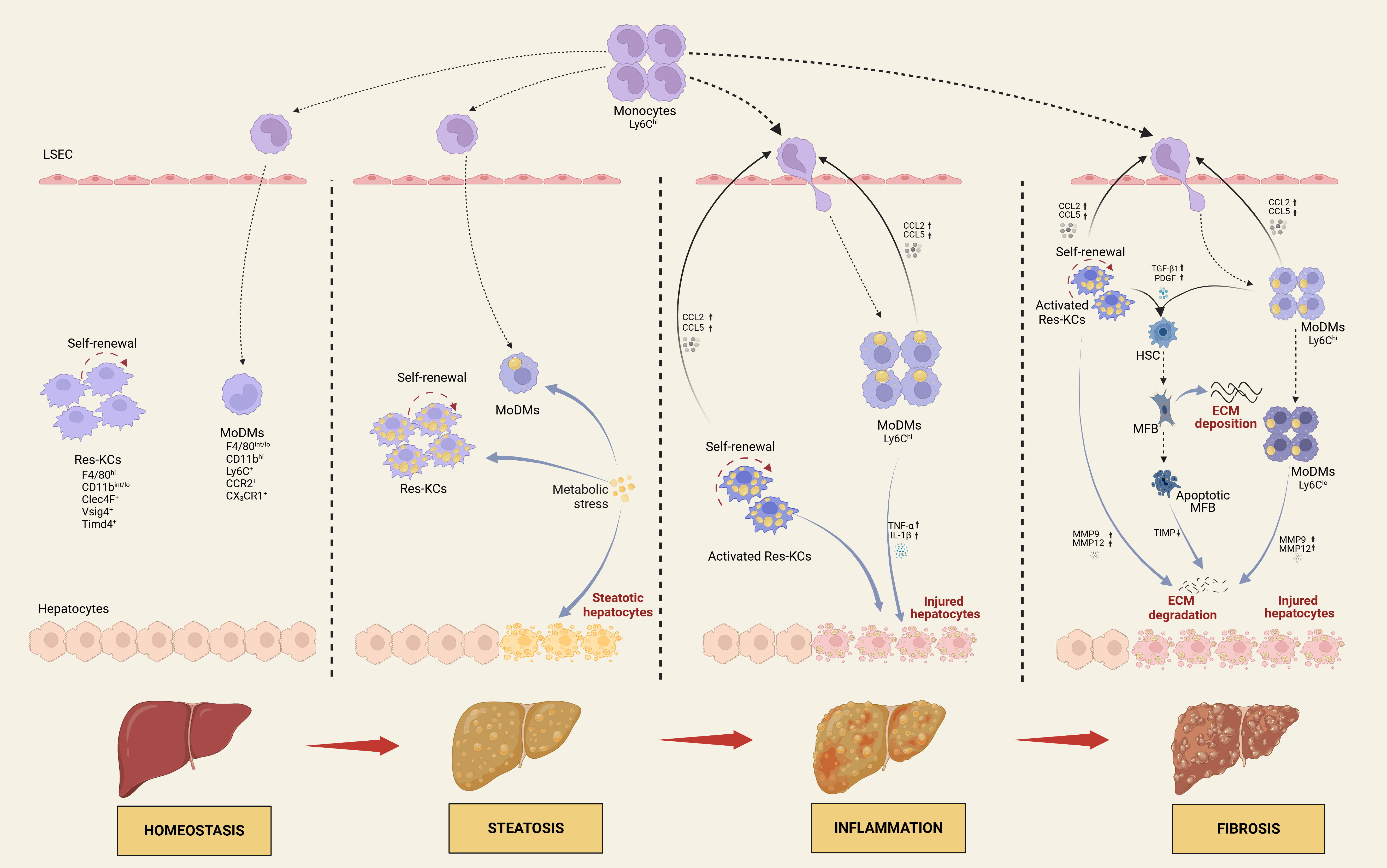
Keywords
metabolic dysfunction-associated steatotic liver disease | macrophage | resident Kupffer cell | monocyte-derived macrophage | therapy
References
- 1.Quek, J.; Chan, K.E.; Wong, Z.Y.; Tan, C.; Tan, B.; Lim, W.H.; Tan, D.J.H.; Tang, A.S.P.; Tay, P.; Xiao, J.; et al. Global prevalence of non-alcoholic fatty liver disease and non-alcoholic steatohepatitis in the overweight and obese population: A systematic review and meta-analysis. Lancet Gastroenterol. Hepatol. 2023, 8, 20–30. https://doi.org/10.1016/s2468-1253(22)00317-x.
- 2.Younossi, Z.M.; Koenig, A.B.; Abdelatif, D.; Fazel, Y.; Henry, L.; Wymer, M. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology 2016, 64, 73–84. https://doi.org/10.1002/hep.28431.
- 3.Riazi, K.; Azhari, H.; Charette, J.H.; Underwood, F.E.; King, J.A.; Afshar, E.E.; Swain, M.G.; Congly, S.E.; Kaplan, G.G.; Shaheen, A.A. The prevalence and incidence of NAFLD worldwide: A systematic review and meta-analysis. Lancet Gastroenterol. Hepatol. 2022, 7, 851–861. https://doi.org/10.1016/s2468-1253(22)00165-0.
- 4.Younossi, Z.M.; Golabi, P.; Paik, J.M.; Henry, A.; Van Dongen, C.; Henry, L. The global epidemiology of nonalcoholic fatty liver disease (NAFLD) and nonalcoholic steatohepatitis (NASH): A systematic review. Hepatology 2023, 77, 1335–1347. https://doi.org/10.1097/hep.0000000000000004.
- 5.Semmler, G.; Datz, C.; Reiberger, T.; Trauner, M. Diet and exercise in NAFLD/NASH: Beyond the obvious. Liver Int. 2021, 41, 2249–2268. https://doi.org/10.1111/liv.15024.
- 6.Wang, X.; He, Q.; Zhou, C.; Xu, Y.; Liu, D.; Fujiwara, N.; Kubota, N.; Click, A.; Henderson, P.; Vancil, J.; et al. Prolonged hypernutrition impairs TREM2-dependent efferocytosis to license chronic liver inflammation and NASH development. Immunity 2023, 56, 58–77.e11. https://doi.org/10.1016/j.immuni.2022.11.013.
- 7.Rinella, M.E.; Lazarus, J.V.; Ratziu, V.; Francque, S.M.; Sanyal, A.J.; Kanwal, F.; Romero, D.; Abdelmalek, M.F.; Anstee, Q.M.; Arab, J.P.; et al. A multisociety Delphi consensus statement on new fatty liver disease nomenclature. J. Hepatol. 2023, 79, 1542–1556. https://doi.org/10.1016/j.jhep.2023.06.003.
- 8.Rinella, M.E.; Neuschwander-Tetri, B.A.; Siddiqui, M.S.; Abdelmalek, M.F.; Caldwell, S.; Barb, D.; Kleiner, D.E.; Loomba, R. AASLD Practice Guidance on the clinical assessment and management of nonalcoholic fatty liver disease. Hepatology 2023, 77, 1797–1835. https://doi.org/10.1097/hep.0000000000000323.
- 9.Hagström, H.; Vessby, J.; Ekstedt, M.; Shang, Y. 99% of patients with NAFLD meet MASLD criteria and natural history is therefore identical. J. Hepatol. 2024, 80, e76–e77. https://doi.org/10.1016/j.jhep.2023.08.026.
- 10.Song, S.J.; Lai, J.C.; Wong, G.L.; Wong, V.W.; Yip, T.C. Can we use old NAFLD data under the new MASLD definition? J. Hepatol. 2024, 80, e54–e56. https://doi.org/10.1016/j.jhep.2023.07.021.
- 11.Campana, L.; Esser, H.; Huch, M.; Forbes, S. Liver regeneration and inflammation: From fundamental science to clinical applications. Nat. Rev. Mol. Cell Biol. 2021, 22, 608–624. https://doi.org/10.1038/s41580-021-00373-7.
- 12.Wang, X.; Wang, Z.; Liu, B.; Jin, R.; Song, Y.; Fei, R.; Cong, X.; Huang, R.; Li, X.; Yang, J.; et al. Characteristic gene expression in the liver monocyte-macrophage-DC system is associated with the progression of fibrosis in NASH. Front. Immunol. 2023, 14, 1098056. https://doi.org/10.3389/fimmu.2023.1098056.
- 13.Guilliams, M.; Dutertre, C.A.; Scott, C.L.; McGovern, N.; Sichien, D.; Chakarov, S.; Van Gassen, S.; Chen, J.; Poidinger, M.; De Prijck, S.; et al. Unsupervised High-Dimensional Analysis Aligns Dendritic Cells across Tissues and Species. Immunity 2016, 45, 669–684. https://doi.org/10.1016/j.immuni.2016.08.015.
- 14.Deppermann, C.; Kratofil, R.M.; Peiseler, M.; David, B.A.; Zindel, J.; Castanheira, F.; van der Wal, F.; Carestia, A.; Jenne, C.N.; Marth, J.D.; et al. Macrophage galactose lectin is critical for Kupffer cells to clear aged platelets. J. Exp. Med. 2020, 217, e20190723. https://doi.org/10.1084/jem.20190723.
- 15.You, Q.; Cheng, L.; Kedl, R.M.; Ju, C. Mechanism of T cell tolerance induction by murine hepatic Kupffer cells. Hepatology 2008, 48, 978–990. https://doi.org/10.1002/hep.22395.
- 16.Wang, Y.; van der Tuin, S.; Tjeerdema, N.; van Dam, A.D.; Rensen, S.S.; Hendrikx, T.; Berbée, J.F.; Atanasovska, B.; Fu, J.; Hoekstra, M.; et al. Plasma cholesteryl ester transfer protein is predominantly derived from Kupffer cells. Hepatology 2015, 62, 1710–1722. https://doi.org/10.1002/hep.27985.
- 17.Guilliams, M.; Scott, C.L. Does niche competition determine the origin of tissue-resident macrophages? Nat. Rev. Immunol. 2017, 17, 451–460. https://doi.org/10.1038/nri.2017.42.
- 18.Krenkel, O.; Tacke, F. Liver macrophages in tissue homeostasis and disease. Nat. Rev. Immunol. 2017, 17, 306–321. https://doi.org/10.1038/nri.2017.11.
- 19.Dou, L.; Shi, X.; He, X.; Gao, Y. Macrophage Phenotype and Function in Liver Disorder. Front. Immunol. 2019, 10, 3112. https://doi.org/10.3389/fimmu.2019.03112.
- 20.Seidman, J.S.; Troutman, T.D.; Sakai, M.; Gola, A.; Spann, N.J.; Bennett, H.; Bruni, C.M.; Ouyang, Z.; Li, R.Z.; Sun, X.; et al. Niche-Specific Reprogramming of Epigenetic Landscapes Drives Myeloid Cell Diversity in Nonalcoholic Steatohepatitis. Immunity 2020, 52, 1057-1074.e1057. https://doi.org/10.1016/j.immuni.2020.04.001.
- 21.MacParland, S.A.; Liu, J.C.; Ma, X.Z.; Innes, B.T.; Bartczak, A.M.; Gage, B.K.; Manuel, J.; Khuu, N.; Echeverri, J.; Linares, I.; et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat. Commun. 2018, 9, 4383. https://doi.org/10.1038/s41467-018-06318-7.
- 22.Schulz, C.; Gomez Perdiguero, E.; Chorro, L.; Szabo-Rogers, H.; Cagnard, N.; Kierdorf, K.; Prinz, M.; Wu, B.; Jacobsen, S.E.; Pollard, J.W.; et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science 2012, 336, 86–90. https://doi.org/10.1126/science.1219179.
- 23.Hoeffel, G.; Chen, J.; Lavin, Y.; Low, D.; Almeida, F.F.; See, P.; Beaudin, A.E.; Lum, J.; Low, I.; Forsberg, E.C.; et al. C-Myb(+) erythro-myeloid progenitor-derived fetal monocytes give rise to adult tissue-resident macrophages. Immunity 2015, 42, 665–678. https://doi.org/10.1016/j.immuni.2015.03.011.
- 24.Zhao, Y.; Zou, W.; Du, J.; Zhao, Y. The origins and homeostasis of monocytes and tissue-resident macrophages in physiological situation. J. Cell Physiol. 2018, 233, 6425–6439. https://doi.org/10.1002/jcp.26461.
- 25.Mass, E.; Ballesteros, I.; Farlik, M.; Halbritter, F.; Günther, P.; Crozet, L.; Jacome-Galarza, C.E.; Händler, K.; Klughammer, J.; Kobayashi, Y.; et al. Specification of tissue-resident macrophages during organogenesis. Science 2016, 353, aaf4238. https://doi.org/10.1126/science.aaf4238.
- 26.Matsuoka, S.; Tsuji, K.; Hisakawa, H.; Xu, M.; Ebihara, Y.; Ishii, T.; Sugiyama, D.; Manabe, A.; Tanaka, R.; Ikeda, Y.; et al. Generation of definitive hematopoietic stem cells from murine early yolk sac and paraaortic splanchnopleures by aorta-gonad-mesonephros region-derived stromal cells. Blood 2001, 98, 6–12. https://doi.org/10.1182/blood.v98.1.6.
- 27.Cox, N.; Pokrovskii, M.; Vicario, R.; Geissmann, F. Origins, Biology, and Diseases of Tissue Macrophages. Annu. Rev. Immunol. 2021, 39, 313–344. https://doi.org/10.1146/annurev-immunol-093019-111748.
- 28.Ginhoux, F.; Jung, S. Monocytes and macrophages: Developmental pathways and tissue homeostasis. Nat. Rev. Immunol. 2014, 14, 392–404. https://doi.org/10.1038/nri3671.
- 29.Remmerie, A.; Martens, L.; Thoné, T.; Castoldi, A.; Seurinck, R.; Pavie, B.; Roels, J.; Vanneste, B.; De Prijck, S.; Vanhockerhout, M.; et al. Osteopontin Expression Identifies a Subset of Recruited Macrophages Distinct from Kupffer Cells in the Fatty Liver. Immunity 2020, 53, 641–657.e14. https://doi.org/10.1016/j.immuni.2020.08.004.
- 30.Liu, Z.; Gu, Y.; Chakarov, S.; Bleriot, C.; Kwok, I.; Chen, X.; Shin, A.; Huang, W.; Dress, R.J.; Dutertre, C.A.; et al. Fate Mapping via Ms4a3-Expression History Traces Monocyte-Derived Cells. Cell 2019, 178, 1509-1525.e1519. https://doi.org/10.1016/j.cell.2019.08.009.
- 31.Chen, J.; Deng, X.; Liu, Y.; Tan, Q.; Huang, G.; Che, Q.; Guo, J.; Su, Z. Kupffer Cells in Non-alcoholic Fatty Liver Disease: Friend or Foe? Int. J. Biol. Sci. 2020, 16, 2367–2378. https://doi.org/10.7150/ijbs.47143.
- 32.Bonnardel, J.; T'Jonck, W.; Gaublomme, D.; Browaeys, R.; Scott, C.L.; Martens, L.; Vanneste, B.; De Prijck, S.; Nedospasov, S.A.; Kremer, A.; et al. Stellate Cells, Hepatocytes, and Endothelial Cells Imprint the Kupffer Cell Identity on Monocytes Colonizing the Liver Macrophage Niche. Immunity 2019, 51, 638–654.e9. https://doi.org/10.1016/j.immuni.2019.08.017.
- 33.Guilliams, M.; Bonnardel, J.; Haest, B.; Vanderborght, B.; Wagner, C.; Remmerie, A.; Bujko, A.; Martens, L.; Thoné, T.; Browaeys, R.; et al. Spatial proteogenomics reveals distinct and evolutionarily conserved hepatic macrophage niches. Cell 2022, 185, 379-396.e338. https://doi.org/10.1016/j.cell.2021.12.018.
- 34.Lavin, Y.; Winter, D.; Blecher-Gonen, R.; David, E.; Keren-Shaul, H.; Merad, M.; Jung, S.; Amit, I. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell 2014, 159, 1312–1326. https://doi.org/10.1016/j.cell.2014.11.018.
- 35.Austyn, J.M.; Gordon, S. F4/80, a monoclonal antibody directed specifically against the mouse macrophage. Eur. J. Immunol. 1981, 11, 805–815. https://doi.org/10.1002/eji.1830111013.
- 36.Ouyang, Z.; Felix, J.; Zhou, J.; Pei, Y.; Ma, B.; Hwang, P.M.; Lemieux, M.J.; Gutsche, I.; Zheng, F.; Wen, Y. Trimeric structure of the mouse Kupffer cell C-type lectin receptor Clec4f. FEBS Lett. 2020, 594, 189–198. https://doi.org/10.1002/1873-3468.13565.
- 37.Vogt, L.; Schmitz, N.; Kurrer, M.O.; Bauer, M.; Hinton, H.I.; Behnke, S.; Gatto, D.; Sebbel, P.; Beerli, R.R.; Sonderegger, I.; et al. VSIG4, a B7 family-related protein, is a negative regulator of T cell activation. J. Clin. Invest. 2006, 116, 2817–2826. https://doi.org/10.1172/jci25673.
- 38.Helmy, K.Y.; Katschke, K.J., Jr.; Gorgani, N.N.; Kljavin, N.M.; Elliott, J.M.; Diehl, L.; Scales, S.J.; Ghilardi, N.; van Lookeren Campagne, M. CRIg: A macrophage complement receptor required for phagocytosis of circulating pathogens. Cell 2006, 124, 915–927. https://doi.org/10.1016/j.cell.2005.12.039.
- 39.Dick, S.A.; Macklin, J.A.; Nejat, S.; Momen, A.; Clemente-Casares, X.; Althagafi, M.G.; Chen, J.; Kantores, C.; Hosseinzadeh, S.; Aronoff, L.; et al. Self-renewing resident cardiac macrophages limit adverse remodeling following myocardial infarction. Nat. Immunol. 2019, 20, 29–39. https://doi.org/10.1038/s41590-018-0272-2.
- 40.Karlmark, K.R.; Weiskirchen, R.; Zimmermann, H.W.; Gassler, N.; Ginhoux, F.; Weber, C.; Merad, M.; Luedde, T.; Trautwein, C.; Tacke, F. Hepatic recruitment of the inflammatory Gr1+ monocyte subset upon liver injury promotes hepatic fibrosis. Hepatology 2009, 50, 261–274. https://doi.org/10.1002/hep.22950.
- 41.Soysa, R.; Lampert, S.; Yuen, S.; Douglass, A.N.; Li, W.; Pfeffer, K.; Crispe, I.N. Fetal origin confers radioresistance on liver macrophages via p21(cip1/WAF1). J. Hepatol. 2019, 71, 553–562. https://doi.org/10.1016/j.jhep.2019.04.015.
- 42.Blériot, C.; Barreby, E.; Dunsmore, G.; Ballaire, R.; Chakarov, S.; Ficht, X.; De Simone, G.; Andreata, F.; Fumagalli, V.; Guo, W.; et al. A subset of Kupffer cells regulates metabolism through the expression of CD36. Immunity 2021, 54, 2101–2116.e6. https://doi.org/10.1016/j.immuni.2021.08.006.
- 43.Krenkel, O.; Hundertmark, J.; Abdallah, A.T.; Kohlhepp, M.; Puengel, T.; Roth, T.; Branco, D.P.P.; Mossanen, J.C.; Luedde, T.; Trautwein, C.; et al. Myeloid cells in liver and bone marrow acquire a functionally distinct inflammatory phenotype during obesity-related steatohepatitis. Gut 2020, 69, 551–563. https://doi.org/10.1136/gutjnl-2019-318382.
- 44.Scott, C.L.; Zheng, F.; De Baetselier, P.; Martens, L.; Saeys, Y.; De Prijck, S.; Lippens, S.; Abels, C.; Schoonooghe, S.; Raes, G.; et al. Bone marrow-derived monocytes give rise to self-renewing and fully differentiated Kupffer cells. Nat. Commun. 2016, 7, 10321. https://doi.org/10.1038/ncomms10321.
- 45.Zimmermann, H.W.; Seidler, S.; Gassler, N.; Nattermann, J.; Luedde, T.; Trautwein, C.; Tacke, F. Interleukin-8 is activated in patients with chronic liver diseases and associated with hepatic macrophage accumulation in human liver fibrosis. PLoS ONE 2011, 6, e21381. https://doi.org/10.1371/journal.pone.0021381.
- 46.Liebold, I.; Meyer, S.; Heine, M.; Kuhl, A.; Witt, J.; Eissing, L.; Fischer, A.W.; Koop, A.C.; Kluwe, J.; Wiesch, J.S.Z.; et al. TREM2 Regulates the Removal of Apoptotic Cells and Inflammatory Processes during the Progression of NAFLD. Cells 2023, 12, 341. https://doi.org/10.3390/cells12030341.
- 47.Gola, A.; Dorrington, M.G.; Speranza, E.; Sala, C.; Shih, R.M.; Radtke, A.J.; Wong, H.S.; Baptista, A.P.; Hernandez, J.M.; Castellani, G.; et al. Commensal-driven immune zonation of the liver promotes host defence. Nature 2021, 589, 131–136. https://doi.org/10.1038/s41586-020-2977-2.
- 48.Dixon, L.J.; Barnes, M.; Tang, H.; Pritchard, M.T.; Nagy, L.E. Kupffer cells in the liver. Compr. Physiol. 2013, 3, 785–797. https://doi.org/10.1002/cphy.c120026.
- 49.Lee, S.J.; Park, S.Y.; Jung, M.Y.; Bae, S.M.; Kim, I.S. Mechanism for phosphatidylserine-dependent erythrophagocytosis in mouse liver. Blood 2011, 117, 5215–5223. https://doi.org/10.1182/blood-2010-10-313239.
- 50.Heymann, F.; Peusquens, J.; Ludwig-Portugall, I.; Kohlhepp, M.; Ergen, C.; Niemietz, P.; Martin, C.; van Rooijen, N.; Ochando, J.C.; Randolph, G.J.; et al. Liver inflammation abrogates immunological tolerance induced by Kupffer cells. Hepatology 2015, 62, 279–291. https://doi.org/10.1002/hep.27793.
- 51.Lombardi, R.; Piciotti, R.; Dongiovanni, P.; Meroni, M.; Fargion, S.; Fracanzani, A.L. PD-1/PD-L1 Immuno-Mediated Therapy in NAFLD: Advantages and Obstacles in the Treatment of Advanced Disease. Int. J. Mol. Sci. 2022, 23, 2707. https://doi.org/10.3390/ijms23052707.
- 52.Li, W.; Yang, Y.; Yang, L.; Chang, N.; Li, L. Monocyte-derived Kupffer cells dominate in the Kupffer cell pool during liver injury. Cell Rep. 2023, 42, 113164. https://doi.org/10.1016/j.celrep.2023.113164.
- 53.Wan, J.; Benkdane, M.; Teixeira-Clerc, F.; Bonnafous, S.; Louvet, A.; Lafdil, F.; Pecker, F.; Tran, A.; Gual, P.; Mallat, A.; et al. M2 Kupffer cells promote M1 Kupffer cell apoptosis: A protective mechanism against alcoholic and nonalcoholic fatty liver disease. Hepatology 2014, 59, 130–142. https://doi.org/10.1002/hep.26607.
- 54.Han, M.; Geng, J.; Zhang, S.; Rao, J.; Zhu, Y.; Xu, S.; Wang, F.; Ma, F.; Zhou, M.; Zhou, H. Invariant natural killer T cells drive hepatic homeostasis in nonalcoholic fatty liver disease via sustained IL-10 expression in CD170(+) Kupffer cells. Eur. J. Immunol. 2023, 53, e2350474. https://doi.org/10.1002/eji.202350474.
- 55.van der Tuin, S.J.L.; Li, Z.; Berbée, J.F.P.; Verkouter, I.; Ringnalda, L.E.; Neele, A.E.; van Klinken, J.B.; Rensen, S.S.; Fu, J.; de Winther, M.P.J.; et al. Lipopolysaccharide Lowers Cholesteryl Ester Transfer Protein by Activating F4/80(+)Clec4f(+)Vsig4(+)Ly6C(-) Kupffer Cell Subsets. J. Am. Heart Assoc. 2018, 7, e008105. https://doi.org/10.1161/jaha.117.008105.
- 56.Naik, S.U.; Wang, X.; Da Silva, J.S.; Jaye, M.; Macphee, C.H.; Reilly, M.P.; Billheimer, J.T.; Rothblat, G.H.; Rader, D.J. Pharmacological activation of liver X receptors promotes reverse cholesterol transport in vivo. Circulation 2006, 113, 90–97. https://doi.org/10.1161/circulationaha.105.560177.
- 57.Theurl, I.; Hilgendorf, I.; Nairz, M.; Tymoszuk, P.; Haschka, D.; Asshoff, M.; He, S.; Gerhardt, L.M.; Holderried, T.A.; Seifert, M.; et al. On-demand erythrocyte disposal and iron recycling requires transient macrophages in the liver. Nat. Med. 2016, 22, 945–951. https://doi.org/10.1038/nm.4146.
- 58.Terpstra, V.; van Berkel, T.J. Scavenger receptors on liver Kupffer cells mediate the in vivo uptake of oxidatively damaged red blood cells in mice. Blood 2000, 95, 2157–2163.
- 59.Theurl, M.; Theurl, I.; Hochegger, K.; Obrist, P.; Subramaniam, N.; van Rooijen, N.; Schuemann, K.; Weiss, G. Kupffer cells modulate iron homeostasis in mice via regulation of hepcidin expression. J. Mol. Med. 2008, 86, 825–835. https://doi.org/10.1007/s00109-008-0346-y.
- 60.Jiang, S.; Yan, K.; Sun, B.; Gao, S.; Yang, X.; Ni, Y.; Ma, W.; Zhao, R. Long-Term High-Fat Diet Decreases Hepatic Iron Storage Associated with Suppressing TFR2 and ZIP14 Expression in Rats. J. Agric. Food Chem. 2018, 66, 11612–11621. https://doi.org/10.1021/acs.jafc.8b02974.
- 61.Tran, S.; Baba, I.; Poupel, L.; Dussaud, S.; Moreau, M.; Gélineau, A.; Marcelin, G.; Magréau-Davy, E.; Ouhachi, M.; Lesnik, P.; et al. Impaired Kupffer Cell Self-Renewal Alters the Liver Response to Lipid Overload during Non-alcoholic Steatohepatitis. Immunity 2020, 53, 627–640.e5. https://doi.org/10.1016/j.immuni.2020.06.003.
- 62.McGettigan, B.; McMahan, R.; Orlicky, D.; Burchill, M.; Danhorn, T.; Francis, P.; Cheng, L.L.; Golden-Mason, L.; Jakubzick, C.V.; Rosen, H.R. Dietary Lipids Differentially Shape Nonalcoholic Steatohepatitis Progression and the Transcriptome of Kupffer Cells and Infiltrating Macrophages. Hepatology 2019, 70, 67–83. https://doi.org/10.1002/hep.30401.
- 63.Sierro, F.; Evrard, M.; Rizzetto, S.; Melino, M.; Mitchell, A.J.; Florido, M.; Beattie, L.; Walters, S.B.; Tay, S.S.; Lu, B.; et al. A Liver Capsular Network of Monocyte-Derived Macrophages Restricts Hepatic Dissemination of Intraperitoneal Bacteria by Neutrophil Recruitment. Immunity 2017, 47, 374-388.e376. https://doi.org/10.1016/j.immuni.2017.07.018.
- 64.Blériot, C.; Dupuis, T.; Jouvion, G.; Eberl, G.; Disson, O.; Lecuit, M. Liver-resident macrophage necroptosis orchestrates type 1 microbicidal inflammation and type-2-mediated tissue repair during bacterial infection. Immunity 2015, 42, 145–158. https://doi.org/10.1016/j.immuni.2014.12.020.
- 65.Donnelly, K.L.; Smith, C.I.; Schwarzenberg, S.J.; Jessurun, J.; Boldt, M.D.; Parks, E.J. Sources of fatty acids stored in liver and secreted via lipoproteins in patients with nonalcoholic fatty liver disease. J. Clin. Invest. 2005, 115, 1343–1351. https://doi.org/10.1172/jci23621.
- 66.Lima, M.L.; Leite, L.H.; Gioda, C.R.; Leme, F.O.; Couto, C.A.; Coimbra, C.C.; Leite, V.H.; Ferrari, T.C. A Novel Wistar Rat Model of Obesity-Related Nonalcoholic Fatty Liver Disease Induced by Sucrose-Rich Diet. J. Diabetes Res. 2016, 2016, 9127076. https://doi.org/10.1155/2016/9127076.
- 67.Bieghs, V.; Wouters, K.; van Gorp, P.J.; Gijbels, M.J.; de Winther, M.P.; Binder, C.J.; Lütjohann, D.; Febbraio, M.; Moore, K.J.; van Bilsen, M.; et al. Role of scavenger receptor A and CD36 in diet-induced nonalcoholic steatohepatitis in hyperlipidemic mice. Gastroenterology 2010, 138, 2477–2486, 2486.e2471-2473. https://doi.org/10.1053/j.gastro.2010.02.051.
- 68.Govaere, O.; Petersen, S.K.; Martinez-Lopez, N.; Wouters, J.; Van Haele, M.; Mancina, R.M.; Jamialahmadi, O.; Bilkei-Gorzo, O.; Lassen, P.B.; Darlay, R.; et al. Macrophage scavenger receptor 1 mediates lipid-induced inflammation in non-alcoholic fatty liver disease. J. Hepatol. 2022, 76, 1001–1012. https://doi.org/10.1016/j.jhep.2021.12.012.
- 69.Leroux, A.; Ferrere, G.; Godie, V.; Cailleux, F.; Renoud, M.L.; Gaudin, F.; Naveau, S.; Prévot, S.; Makhzami, S.; Perlemuter, G.; et al. Toxic lipids stored by Kupffer cells correlates with their pro-inflammatory phenotype at an early stage of steatohepatitis. J. Hepatol. 2012, 57, 141–149. https://doi.org/10.1016/j.jhep.2012.02.028.
- 70.Huang, W.; Metlakunta, A.; Dedousis, N.; Zhang, P.; Sipula, I.; Dube, J.J.; Scott, D.K.; O’Doherty, R.M. Depletion of liver Kupffer cells prevents the development of diet-induced hepatic steatosis and insulin resistance. Diabetes 2010, 59, 347–357. https://doi.org/10.2337/db09-0016.
- 71.Stienstra, R.; Saudale, F.; Duval, C.; Keshtkar, S.; Groener, J.E.; van Rooijen, N.; Staels, B.; Kersten, S.; Müller, M. Kupffer cells promote hepatic steatosis via interleukin-1beta-dependent suppression of peroxisome proliferator-activated receptor alpha activity. Hepatology 2010, 51, 511–522. https://doi.org/10.1002/hep.23337.
- 72.Iwata, H.; Osborn, E.A.; Ughi, G.J.; Murakami, K.; Goettsch, C.; Hutcheson, J.D.; Mauskapf, A.; Mattson, P.C.; Libby, P.; Singh, S.A.; et al. Highly Selective PPARα (Peroxisome Proliferator-Activated Receptor α) Agonist Pemafibrate Inhibits Stent Inflammation and Restenosis Assessed by Multimodality Molecular-Microstructural Imaging. J. Am. Heart Assoc. 2021, 10, e020834. https://doi.org/10.1161/jaha.121.020834.
- 73.Clementi, A.H.; Gaudy, A.M.; van Rooijen, N.; Pierce, R.H.; Mooney, R.A. Loss of Kupffer cells in diet-induced obesity is associated with increased hepatic steatosis, STAT3 signaling, and further decreases in insulin signaling. Biochim. Biophys. Acta 2009, 1792, 1062–1072. https://doi.org/10.1016/j.bbadis.2009.08.007.
- 74.Gao, H.; Jin, Z.; Bandyopadhyay, G.; Cunha, E.R.K.; Liu, X.; Zhao, H.; Zhang, D.; Jouihan, H.; Pourshahian, S.; Kisseleva, T.; et al. MiR-690 treatment causes decreased fibrosis and steatosis and restores specific Kupffer cell functions in NASH. Cell Metab. 2022, 34, 978-990.e974. https://doi.org/10.1016/j.cmet.2022.05.008.
- 75.Zhong, L.; Xu, K.S.; Deng, L. Study on the state of macrophage infiltration in the progression of non-alcoholic fatty liver disease induced by high-fat diet in mice. Zhonghua Gan Zang Bing. Za Zhi 2020, 28, 1042–1047. https://doi.org/10.3760/cma.j.cn501113-20190712-00244.
- 76.Barreby, E.; Chen, P.; Aouadi, M. Macrophage functional diversity in NAFLD—More than inflammation. Nat. Rev. Endocrinol. 2022, 18, 461–472. https://doi.org/10.1038/s41574-022-00675-6.
- 77.Westerbacka, J.; Kolak, M.; Kiviluoto, T.; Arkkila, P.; Sirén, J.; Hamsten, A.; Fisher, R.M.; Yki-Järvinen, H. Genes involved in fatty acid partitioning and binding, lipolysis, monocyte/macrophage recruitment, and inflammation are overexpressed in the human fatty liver of insulin-resistant subjects. Diabetes 2007, 56, 2759–2765. https://doi.org/10.2337/db07-0156.
- 78.Gadd, V.L.; Skoien, R.; Powell, E.E.; Fagan, K.J.; Winterford, C.; Horsfall, L.; Irvine, K.; Clouston, A.D. The portal inflammatory infiltrate and ductular reaction in human nonalcoholic fatty liver disease. Hepatology 2014, 59, 1393–1405. https://doi.org/10.1002/hep.26937.
- 79.Yamaguchi, K.; Yang, L.; McCall, S.; Huang, J.; Yu, X.X.; Pandey, S.K.; Bhanot, S.; Monia, B.P.; Li, Y.X.; Diehl, A.M. Inhibiting triglyceride synthesis improves hepatic steatosis but exacerbates liver damage and fibrosis in obese mice with nonalcoholic steatohepatitis. Hepatology 2007, 45, 1366–1374. https://doi.org/10.1002/hep.21655.
- 80.Teli, M.R.; James, O.F.; Burt, A.D.; Bennett, M.K.; Day, C.P. The natural history of nonalcoholic fatty liver: A follow-up study. Hepatology 1995, 22, 1714–1719.
- 81.Pais, R.; Charlotte, F.; Fedchuk, L.; Bedossa, P.; Lebray, P.; Poynard, T.; Ratziu, V. A systematic review of follow-up biopsies reveals disease progression in patients with non-alcoholic fatty liver. J. Hepatol. 2013, 59, 550–556. https://doi.org/10.1016/j.jhep.2013.04.027.
- 82.Simon, G.; Heckmann, V.; Tóth, D.; Pauka, D.; Petrus, K.; Molnár, T.F. The effect of hepatic steatosis and fibrosis on liver weight and dimensions. Leg. Med. 2020, 47, 101781. https://doi.org/10.1016/j.legalmed.2020.101781.
- 83.Chalasani, N.; Wilson, L.; Kleiner, D.E.; Cummings, O.W.; Brunt, E.M.; Unalp, A. Relationship of steatosis grade and zonal location to histological features of steatohepatitis in adult patients with non-alcoholic fatty liver disease. J. Hepatol. 2008, 48, 829–834. https://doi.org/10.1016/j.jhep.2008.01.016.
- 84.Ramnath, D.; Irvine, K.M.; Lukowski, S.W.; Horsfall, L.U.; Loh, Z.; Clouston, A.D.; Patel, P.J.; Fagan, K.J.; Iyer, A.; Lampe, G.; et al. Hepatic expression profiling identifies steatosis-independent and steatosis-driven advanced fibrosis genes. JCI Insight 2018, 3. https://doi.org/10.1172/jci.insight.120274.
- 85.Chalasani, N.; Younossi, Z.; Lavine, J.E.; Diehl, A.M.; Brunt, E.M.; Cusi, K.; Charlton, M.; Sanyal, A.J. The diagnosis and management of non-alcoholic fatty liver disease: Practice guideline by the American Gastroenterological Association, American Association for the Study of Liver Diseases, and American College of Gastroenterology. Gastroenterology 2012, 142, 1592–1609. https://doi.org/10.1053/j.gastro.2012.04.001.
- 86.Daemen, S.; Gainullina, A.; Kalugotla, G.; He, L.; Chan, M.M.; Beals, J.W.; Liss, K.H.; Klein, S.; Feldstein, A.E.; Finck, B.N.; et al. Dynamic Shifts in the Composition of Resident and Recruited Macrophages Influence Tissue Remodeling in NASH. Cell Rep. 2021, 34, 108626. https://doi.org/10.1016/j.celrep.2020.108626.
- 87.Wu, J.; Li, J.; Salcedo, R.; Mivechi, N.F.; Trinchieri, G.; Horuzsko, A. The proinflammatory myeloid cell receptor TREM-1 controls Kupffer cell activation and development of hepatocellular carcinoma. Cancer Res. 2012, 72, 3977–3986. https://doi.org/10.1158/0008-5472.Can-12-0938.
- 88.Orfila, C.; Lepert, J.C.; Alric, L.; Carrera, G.; Beraud, M.; Vinel, J.P.; Pipy, B. Expression of TNF-alpha and immunohistochemical distribution of hepatic macrophage surface markers in carbon tetrachloride-induced chronic liver injury in rats. Histochem. J. 1999, 31, 677–685. https://doi.org/10.1023/a:1003851821487.
- 89.Kiagiadaki, F.; Kampa, M.; Voumvouraki, A.; Castanas, E.; Kouroumalis, E.; Notas, G. Activin-A causes Hepatic stellate cell activation via the induction of TNFα and TGFβ in Kupffer cells. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 891–899. https://doi.org/10.1016/j.bbadis.2017.12.031.
- 90.Morinaga, H.; Mayoral, R.; Heinrichsdorff, J.; Osborn, O.; Franck, N.; Hah, N.; Walenta, E.; Bandyopadhyay, G.; Pessentheiner, A.R.; Chi, T.J.; et al. Characterization of distinct subpopulations of hepatic macrophages in HFD/obese mice. Diabetes 2015, 64, 1120–1130. https://doi.org/10.2337/db14-1238.
- 91.Kang, J.; Postigo-Fernandez, J.; Kim, K.; Zhu, C.; Yu, J.; Meroni, M.; Mayfield, B.; Bartolomé, A.; Dapito, D.H.; Ferrante, A.W., Jr.; et al. Notch-mediated hepatocyte MCP-1 secretion causes liver fibrosis. JCI Insight 2023, 8, e165369. https://doi.org/10.1172/jci.insight.165369.
- 92.Miura, K.; Yang, L.; van Rooijen, N.; Ohnishi, H.; Seki, E. Hepatic recruitment of macrophages promotes nonalcoholic steatohepatitis through CCR2. Am. J. Physiol. Gastrointest. Liver Physiol. 2012, 302, G1310–G1321. https://doi.org/10.1152/ajpgi.00365.2011.
- 93.Krenkel, O.; Puengel, T.; Govaere, O.; Abdallah, A.T.; Mossanen, J.C.; Kohlhepp, M.; Liepelt, A.; Lefebvre, E.; Luedde, T.; Hellerbrand, C.; et al. Therapeutic inhibition of inflammatory monocyte recruitment reduces steatohepatitis and liver fibrosis. Hepatology 2018, 67, 1270–1283. https://doi.org/10.1002/hep.29544.
- 94.Hendrikx, T.; Porsch, F.; Kiss, M.G.; Rajcic, D.; Papac-Miličević, N.; Hoebinger, C.; Goederle, L.; Hladik, A.; Shaw, L.E.; Horstmann, H.; et al. Soluble TREM2 levels reflect the recruitment and expansion of TREM2(+) macrophages that localize to fibrotic areas and limit NASH. J. Hepatol. 2022, 77, 1373–1385. https://doi.org/10.1016/j.jhep.2022.06.004.
- 95.Tosello-Trampont, A.C.; Landes, S.G.; Nguyen, V.; Novobrantseva, T.I.; Hahn, Y.S. Kuppfer cells trigger nonalcoholic steatohepatitis development in diet-induced mouse model through tumor necrosis factor-α production. J. Biol. Chem. 2012, 287, 40161–40172. https://doi.org/10.1074/jbc.M112.417014.
- 96.Sakai, M.; Troutman, T.D.; Seidman, J.S.; Ouyang, Z.; Spann, N.J.; Abe, Y.; Ego, K.M.; Bruni, C.M.; Deng, Z.; Schlachetzki, J.C.M.; et al. Liver-Derived Signals Sequentially Reprogram Myeloid Enhancers to Initiate and Maintain Kupffer Cell Identity. Immunity 2019, 51, 655–670.e8. https://doi.org/10.1016/j.immuni.2019.09.002.
- 97.Zhang, X.; Fan, L.; Wu, J.; Xu, H.; Leung, W.Y.; Fu, K.; Wu, J.; Liu, K.; Man, K.; Yang, X.; et al. Macrophage p38α promotes nutritional steatohepatitis through M1 polarization. J. Hepatol. 2019, 71, 163–174. https://doi.org/10.1016/j.jhep.2019.03.014.
- 98.Lan, T.; Jiang, S.; Zhang, J.; Weng, Q.; Yu, Y.; Li, H.; Tian, S.; Ding, X.; Hu, S.; Yang, Y.; et al. Breviscapine alleviates NASH by inhibiting TGF-β-activated kinase 1-dependent signaling. Hepatology 2022, 76, 155–171. https://doi.org/10.1002/hep.32221.
- 99.Dulai, P.S.; Singh, S.; Patel, J.; Soni, M.; Prokop, L.J.; Younossi, Z.; Sebastiani, G.; Ekstedt, M.; Hagstrom, H.; Nasr, P.; et al. Increased risk of mortality by fibrosis stage in nonalcoholic fatty liver disease: Systematic review and meta-analysis. Hepatology 2017, 65, 1557–1565. https://doi.org/10.1002/hep.29085.
- 100.Henderson, N.C.; Rieder, F.; Wynn, T.A. Fibrosis: From mechanisms to medicines. Nature 2020, 587, 555–566. https://doi.org/10.1038/s41586-020-2938-9.
- 101.Marra, F. Hepatic stellate cells and the regulation of liver inflammation. J. Hepatol. 1999, 31, 1120–1130. https://doi.org/10.1016/s0168-8278(99)80327-4.
- 102.Mederacke, I.; Hsu, C.C.; Troeger, J.S.; Huebener, P.; Mu, X.; Dapito, D.H.; Pradere, J.P.; Schwabe, R.F. Fate tracing reveals hepatic stellate cells as dominant contributors to liver fibrosis independent of its aetiology. Nat. Commun. 2013, 4, 2823. https://doi.org/10.1038/ncomms3823.
- 103.Lua, I.; Li, Y.; Zagory, J.A.; Wang, K.S.; French, S.W.; Sévigny, J.; Asahina, K. Characterization of hepatic stellate cells, portal fibroblasts, and mesothelial cells in normal and fibrotic livers. J. Hepatol. 2016, 64, 1137–1146. https://doi.org/10.1016/j.jhep.2016.01.010.
- 104.Russo, F.P.; Alison, M.R.; Bigger, B.W.; Amofah, E.; Florou, A.; Amin, F.; Bou-Gharios, G.; Jeffery, R.; Iredale, J.P.; Forbes, S.J. The bone marrow functionally contributes to liver fibrosis. Gastroenterology 2006, 130, 1807–1821. https://doi.org/10.1053/j.gastro.2006.01.036.
- 105.Duffield, J.S.; Forbes, S.J.; Constandinou, C.M.; Clay, S.; Partolina, M.; Vuthoori, S.; Wu, S.; Lang, R.; Iredale, J.P. Selective depletion of macrophages reveals distinct, opposing roles during liver injury and repair. J. Clin. Invest. 2005, 115, 56–65. https://doi.org/10.1172/jci22675.
- 106.Seki, E.; de Minicis, S.; Inokuchi, S.; Taura, K.; Miyai, K.; van Rooijen, N.; Schwabe, R.F.; Brenner, D.A. CCR2 promotes hepatic fibrosis in mice. Hepatology 2009, 50, 185–197. https://doi.org/10.1002/hep.22952.
- 107.Schwabe, R.F.; Tabas, I.; Pajvani, U.B. Mechanisms of Fibrosis Development in Nonalcoholic Steatohepatitis. Gastroenterology 2020, 158, 1913–1928. https://doi.org/10.1053/j.gastro.2019.11.311.
- 108.Xiang, D.; Zou, J.; Zhu, X.; Chen, X.; Luo, J.; Kong, L.; Zhang, H. Physalin D attenuates hepatic stellate cell activation and liver fibrosis by blocking TGF-β/Smad and YAP signaling. Phytomedicine 2020, 78, 153294. https://doi.org/10.1016/j.phymed.2020.153294.
- 109.Lodyga, M.; Cambridge, E.; Karvonen, H.M.; Pakshir, P.; Wu, B.; Boo, S.; Kiebalo, M.; Kaarteenaho, R.; Glogauer, M.; Kapoor, M.; et al. Cadherin-11-mediated adhesion of macrophages to myofibroblasts establishes a profibrotic niche of active TGF-β. Sci. Signal 2019, 12, eaao3469. https://doi.org/10.1126/scisignal.aao3469.
- 110.Wree, A.; McGeough, M.D.; Inzaugarat, M.E.; Eguchi, A.; Schuster, S.; Johnson, C.D.; Peña, C.A.; Geisler, L.J.; Papouchado, B.G.; Hoffman, H.M.; et al. NLRP3 inflammasome driven liver injury and fibrosis: Roles of IL-17 and TNF in mice. Hepatology 2018, 67, 736–749. https://doi.org/10.1002/hep.29523.
- 111.Mridha, A.R.; Wree, A.; Robertson, A.A.B.; Yeh, M.M.; Johnson, C.D.; Van Rooyen, D.M.; Haczeyni, F.; Teoh, N.C.; Savard, C.; Ioannou, G.N.; et al. NLRP3 inflammasome blockade reduces liver inflammation and fibrosis in experimental NASH in mice. J. Hepatol. 2017, 66, 1037–1046. https://doi.org/10.1016/j.jhep.2017.01.022.
- 112.Pradere, J.P.; Kluwe, J.; De Minicis, S.; Jiao, J.J.; Gwak, G.Y.; Dapito, D.H.; Jang, M.K.; Guenther, N.D.; Mederacke, I.; Friedman, R.; et al. Hepatic macrophages but not dendritic cells contribute to liver fibrosis by promoting the survival of activated hepatic stellate cells in mice. Hepatology 2013, 58, 1461–1473. https://doi.org/10.1002/hep.26429.
- 113.Buonomo, E.L.; Mei, S.; Guinn, S.R.; Leo, I.R.; Peluso, M.J.; Nolan, M.A.; Schildberg, F.A.; Zhao, L.; Lian, C.; Xu, S.; et al. Liver stromal cells restrict macrophage maturation and stromal IL-6 limits the differentiation of cirrhosis-linked macrophages. J. Hepatol. 2022, 76, 1127–1137. https://doi.org/10.1016/j.jhep.2021.12.036.
- 114.Fabre, T.; Barron, A.M.S.; Christensen, S.M.; Asano, S.; Bound, K.; Lech, M.P.; Wadsworth, M.H., 2nd; Chen, X.; Wang, C.; Wang, J.; et al. Identification of a broadly fibrogenic macrophage subset induced by type 3 inflammation. Sci. Immunol. 2023, 8, eadd8945. https://doi.org/10.1126/sciimmunol.add8945.
- 115.Ramachandran, P.; Dobie, R.; Wilson-Kanamori, J.R.; Dora, E.F.; Henderson, B.E.P.; Luu, N.T.; Portman, J.R.; Matchett, K.P.; Brice, M.; Marwick, J.A.; et al. Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature 2019, 575, 512–518. https://doi.org/10.1038/s41586-019-1631-3.
- 116.Ramachandran, P.; Pellicoro, A.; Vernon, M.A.; Boulter, L.; Aucott, R.L.; Ali, A.; Hartland, S.N.; Snowdon, V.K.; Cappon, A.; Gordon-Walker, T.T.; et al. Differential Ly-6C expression identifies the recruited macrophage phenotype, which orchestrates the regression of murine liver fibrosis. Proc. Natl. Acad. Sci. USA 2012, 109, E3186-3195. https://doi.org/10.1073/pnas.1119964109.
- 117.Wang, M.; You, Q.; Lor, K.; Chen, F.; Gao, B.; Ju, C. Chronic alcohol ingestion modulates hepatic macrophage populations and functions in mice. J. Leukoc. Biol. 2014, 96, 657–665. https://doi.org/10.1189/jlb.6A0114-004RR.
- 118.Li, Y.H.; Shen, S.; Shao, T.; Jin, M.T.; Fan, D.D.; Lin, A.F.; Xiang, L.X.; Shao, J.Z. Mesenchymal stem cells attenuate liver fibrosis by targeting Ly6C(hi/lo) macrophages through activating the cytokine-paracrine and apoptotic pathways. Cell Death Discov. 2021, 7, 239. https://doi.org/10.1038/s41420-021-00584-z.
- 119.Pellicoro, A.; Aucott, R.L.; Ramachandran, P.; Robson, A.J.; Fallowfield, J.A.; Snowdon, V.K.; Hartland, S.N.; Vernon, M.; Duffield, J.S.; Benyon, R.C.; et al. Elastin accumulation is regulated at the level of degradation by macrophage metalloelastase (MMP-12) during experimental liver fibrosis. Hepatology 2012, 55, 1965–1975. https://doi.org/10.1002/hep.25567.
- 120.Lichtinghagen, R.; Pietsch, D.; Bantel, H.; Manns, M.P.; Brand, K.; Bahr, M.J. The Enhanced Liver Fibrosis (ELF) score: Normal values, influence factors and proposed cut-off values. J. Hepatol. 2013, 59, 236–242. https://doi.org/10.1016/j.jhep.2013.03.016.
- 121.Atabai, K.; Jame, S.; Azhar, N.; Kuo, A.; Lam, M.; McKleroy, W.; Dehart, G.; Rahman, S.; Xia, D.D.; Melton, A.C.; et al. Mfge8 diminishes the severity of tissue fibrosis in mice by binding and targeting collagen for uptake by macrophages. J. Clin. Invest. 2009, 119, 3713–3722. https://doi.org/10.1172/jci40053.
- 122.Hirsova, P.; Ibrahim, S.H.; Krishnan, A.; Verma, V.K.; Bronk, S.F.; Werneburg, N.W.; Charlton, M.R.; Shah, V.H.; Malhi, H.; Gores, G.J. Lipid-Induced Signaling Causes Release of Inflammatory Extracellular Vesicles From Hepatocytes. Gastroenterology 2016, 150, 956–967. https://doi.org/10.1053/j.gastro.2015.12.037.
- 123.Breuer, D.A.; Pacheco, M.C.; Washington, M.K.; Montgomery, S.A.; Hasty, A.H.; Kennedy, A.J. CD8(+) T cells regulate liver injury in obesity-related nonalcoholic fatty liver disease. Am. J. Physiol. Gastrointest. Liver Physiol. 2020, 318, G211–G224. https://doi.org/10.1152/ajpgi.00040.2019.
- 124.Friedman, S.L.; Ratziu, V.; Harrison, S.A.; Abdelmalek, M.F.; Aithal, G.P.; Caballeria, J.; Francque, S.; Farrell, G.; Kowdley, K.V.; Craxi, A.; et al. A randomized, placebo-controlled trial of cenicriviroc for treatment of nonalcoholic steatohepatitis with fibrosis. Hepatology 2018, 67, 1754–1767. https://doi.org/10.1002/hep.29477.
- 125.Anstee, Q.M.; Neuschwander-Tetri, B.A.; Wai-Sun Wong, V.; Abdelmalek, M.F.; Rodriguez-Araujo, G.; Landgren, H.; Park, G.S.; Bedossa, P.; Alkhouri, N.; Tacke, F.; et al. Cenicriviroc Lacked Efficacy to Treat Liver Fibrosis in Nonalcoholic Steatohepatitis: AURORA Phase III Randomized Study. Clin. Gastroenterol. Hepatol. 2024, 22, 124–134.e1. https://doi.org/10.1016/j.cgh.2023.04.003.
- 126.Harrison, S.A.; Marri, S.R.; Chalasani, N.; Kohli, R.; Aronstein, W.; Thompson, G.A.; Irish, W.; Miles, M.V.; Xanthakos, S.A.; Lawitz, E.; et al. Randomised clinical study: GR-MD-02, a galectin-3 inhibitor, vs. placebo in patients having non-alcoholic steatohepatitis with advanced fibrosis. Aliment. Pharmacol. Ther. 2016, 44, 1183–1198. https://doi.org/10.1111/apt.13816.
- 127.Chalasani, N.; Abdelmalek, M.F.; Garcia-Tsao, G.; Vuppalanchi, R.; Alkhouri, N.; Rinella, M.; Noureddin, M.; Pyko, M.; Shiffman, M.; Sanyal, A.; et al. Effects of Belapectin, an Inhibitor of Galectin-3, in Patients With Nonalcoholic Steatohepatitis With Cirrhosis and Portal Hypertension. Gastroenterology 2020, 158, 1334–1345.e5. https://doi.org/10.1053/j.gastro.2019.11.296.
- 128.Svendsen, P.; Graversen, J.H.; Etzerodt, A.; Hager, H.; Røge, R.; Grønbæk, H.; Christensen, E.I.; Møller, H.J.; Vilstrup, H.; Moestrup, S.K. Antibody-Directed Glucocorticoid Targeting to CD163 in M2-type Macrophages Attenuates Fructose-Induced Liver Inflammatory Changes. Mol. Ther. Methods Clin. Dev. 2017, 4, 50–61. https://doi.org/10.1016/j.omtm.2016.11.004.
- 129.Armstrong, M.J.; Gaunt, P.; Aithal, G.P.; Barton, D.; Hull, D.; Parker, R.; Hazlehurst, J.M.; Guo, K.; Abouda, G.; Aldersley, M.A.; et al. Liraglutide safety and efficacy in patients with non-alcoholic steatohepatitis (LEAN): A multicentre, double-blind, randomised, placebo-controlled phase 2 study. Lancet 2016, 387, 679–690. https://doi.org/10.1016/s0140-6736(15)00803-x.
- 130.Newsome, P.N.; Buchholtz, K.; Cusi, K.; Linder, M.; Okanoue, T.; Ratziu, V.; Sanyal, A.J.; Sejling, A.S.; Harrison, S.A. A Placebo-Controlled Trial of Subcutaneous Semaglutide in Nonalcoholic Steatohepatitis. N. Engl. J. Med. 2021, 384, 1113–1124. https://doi.org/10.1056/NEJMoa2028395.
- 131.Loomba, R.; Lawitz, E.; Mantry, P.S.; Jayakumar, S.; Caldwell, S.H.; Arnold, H.; Diehl, A.M.; Djedjos, C.S.; Han, L.; Myers, R.P.; et al. The ASK1 inhibitor selonsertib in patients with nonalcoholic steatohepatitis: A randomized, phase 2 trial. Hepatology 2018, 67, 549–559. https://doi.org/10.1002/hep.29514.
- 132.Harrison, S.A.; Wong, V.W.; Okanoue, T.; Bzowej, N.; Vuppalanchi, R.; Younes, Z.; Kohli, A.; Sarin, S.; Caldwell, S.H.; Alkhouri, N.; et al. Selonsertib for patients with bridging fibrosis or compensated cirrhosis due to NASH: Results from randomized phase III STELLAR trials. J. Hepatol. 2020, 73, 26–39. https://doi.org/10.1016/j.jhep.2020.02.027.
- 133.Neuschwander-Tetri, B.A.; Loomba, R.; Sanyal, A.J.; Lavine, J.E.; Van Natta, M.L.; Abdelmalek, M.F.; Chalasani, N.; Dasarathy, S.; Diehl, A.M.; Hameed, B.; et al. Farnesoid X nuclear receptor ligand obeticholic acid for non-cirrhotic, non-alcoholic steatohepatitis (FLINT): A multicentre, randomised, placebo-controlled trial. Lancet 2015, 385, 956–965. https://doi.org/10.1016/s0140-6736(14)61933-4.
- 134.Younossi, Z.M.; Ratziu, V.; Loomba, R.; Rinella, M.; Anstee, Q.M.; Goodman, Z.; Bedossa, P.; Geier, A.; Beckebaum, S.; Newsome, P.N.; et al. Obeticholic acid for the treatment of non-alcoholic steatohepatitis: Interim analysis from a multicentre, randomised, placebo-controlled phase 3 trial. Lancet 2019, 394, 2184–2196. https://doi.org/10.1016/s0140-6736(19)33041-7.
- 135.Francque, S.M.; Bedossa, P.; Ratziu, V.; Anstee, Q.M.; Bugianesi, E.; Sanyal, A.J.; Loomba, R.; Harrison, S.A.; Balabanska, R.; Mateva, L.; et al. A Randomized, Controlled Trial of the Pan-PPAR Agonist Lanifibranor in NASH. N. Engl. J. Med. 2021, 385, 1547–1558. https://doi.org/10.1056/NEJMoa2036205.
- 136.Zhou, E.; Ge, X.; Nakashima, H.; Li, R.; van der Zande, H.J.P.; Liu, C.; Li, Z.; Müller, C.; Bracher, F.; Mohammed, Y.; et al. Inhibition of DHCR24 activates LXRα to ameliorate hepatic steatosis and inflammation. EMBO Mol. Med. 2023, 15, e16845. https://doi.org/10.15252/emmm.202216845.
- 137.Pérez-Martínez, L.; Ochoa-Callejero, L.; Rubio-Mediavilla, S.; Narro, J.; Bernardo, I.; Oteo, J.A.; Blanco, J.R. Maraviroc improves hepatic triglyceride content but not inflammation in a murine nonalcoholic fatty liver disease model induced by a chronic exposure to high-fat diet. Transl. Res. 2018, 196, 17–30. https://doi.org/10.1016/j.trsl.2018.01.004.
- 138.Baeck, C.; Wehr, A.; Karlmark, K.R.; Heymann, F.; Vucur, M.; Gassler, N.; Huss, S.; Klussmann, S.; Eulberg, D.; Luedde, T.; et al. Pharmacological inhibition of the chemokine CCL2 (MCP-1) diminishes liver macrophage infiltration and steatohepatitis in chronic hepatic injury. Gut 2012, 61, 416–426. https://doi.org/10.1136/gutjnl-2011-300304.
- 139.Yost, R.; Pasquale, T.R.; Sahloff, E.G. Maraviroc: A coreceptor CCR5 antagonist for management of HIV infection. Am. J. Health Syst. Pharm. 2009, 66, 715–726. https://doi.org/10.2146/ajhp080206.
- 140.Traber, P.G.; Zomer, E. Therapy of experimental NASH and fibrosis with galectin inhibitors. PLoS ONE 2013, 8, e83481. https://doi.org/10.1371/journal.pone.0083481.
- 141.Lin, X.; Xu, Y.; Pan, X.; Xu, J.; Ding, Y.; Sun, X.; Song, X.; Ren, Y.; Shan, P.F. Global, regional, and national burden and trend of diabetes in 195 countries and territories: An analysis from 1990 to 2025. Sci Rep 2020, 10, 14790. https://doi.org/10.1038/s41598-020-71908-9.
- 142.Wang, Y.; Parlevliet, E.T.; Geerling, J.J.; van der Tuin, S.J.; Zhang, H.; Bieghs, V.; Jawad, A.H.; Shiri-Sverdlov, R.; Bot, I.; de Jager, S.C.; et al. Exendin-4 decreases liver inflammation and atherosclerosis development simultaneously by reducing macrophage infiltration. Br. J. Pharmacol. 2014, 171, 723–734. https://doi.org/10.1111/bph.12490.
- 143.Perakakis, N.; Stefanakis, K.; Feigh, M.; Veidal, S.S.; Mantzoros, C.S. Elafibranor and liraglutide improve differentially liver health and metabolism in a mouse model of non-alcoholic steatohepatitis. Liver Int. 2021, 41, 1853–1866. https://doi.org/10.1111/liv.14888.
- 144.Lan, T.; Hu, Y.; Hu, F.; Li, H.; Chen, Y.; Zhang, J.; Yu, Y.; Jiang, S.; Weng, Q.; Tian, S.; et al. Hepatocyte glutathione S-transferase mu 2 prevents non-alcoholic steatohepatitis by suppressing ASK1 signaling. J. Hepatol. 2022, 76, 407–419. https://doi.org/10.1016/j.jhep.2021.09.040.
- 145.Immanuel, C.N.; Teng, B.; Dong, B.; Gordon, E.M.; Kennedy, J.A.; Luellen, C.; Schwingshackl, A.; Cormier, S.A.; Fitzpatrick, E.A.; Waters, C.M. Apoptosis signal-regulating kinase-1 promotes inflammasome priming in macrophages. Am. J. Physiol. Lung Cell Mol. Physiol. 2019, 316, L418-l427. https://doi.org/10.1152/ajplung.00199.2018.
- 146.Puengel, T.; De Vos, S.; Hundertmark, J.; Kohlhepp, M.; Guldiken, N.; Pujuguet, P.; Auberval, M.; Marsais, F.; Shoji, K.F.; Saniere, L.; et al. The Medium-Chain Fatty Acid Receptor GPR84 Mediates Myeloid Cell Infiltration Promoting Steatohepatitis and Fibrosis. J. Clin. Med. 2020, 9, 1140. https://doi.org/10.3390/jcm9041140.
- 147.Yao, J.; Zhou, C.S.; Ma, X.; Fu, B.Q.; Tao, L.S.; Chen, M.; Xu, Y.P. FXR agonist GW4064 alleviates endotoxin-induced hepatic inflammation by repressing macrophage activation. World J. Gastroenterol. 2014, 20, 14430–14441. https://doi.org/10.3748/wjg.v20.i39.14430.
- 148.McMahan, R.H.; Wang, X.X.; Cheng, L.L.; Krisko, T.; Smith, M.; El Kasmi, K.; Pruzanski, M.; Adorini, L.; Golden-Mason, L.; Levi, M.; et al. Bile acid receptor activation modulates hepatic monocyte activity and improves nonalcoholic fatty liver disease. J. Biol. Chem. 2013, 288, 11761–11770. https://doi.org/10.1074/jbc.M112.446575.
- 149.Lefere, S.; Puengel, T.; Hundertmark, J.; Penners, C.; Frank, A.K.; Guillot, A.; de Muynck, K.; Heymann, F.; Adarbes, V.; Defrêne, E.; et al. Differential effects of selective- and pan-PPAR agonists on experimental steatohepatitis and hepatic macrophages(☆). J. Hepatol. 2020, 73, 757–770. https://doi.org/10.1016/j.jhep.2020.04.025.
- 150.Montagner, A.; Polizzi, A.; Fouché, E.; Ducheix, S.; Lippi, Y.; Lasserre, F.; Barquissau, V.; Régnier, M.; Lukowicz, C.; Benhamed, F.; et al. Liver PPARα is crucial for whole-body fatty acid homeostasis and is protective against NAFLD. Gut 2016, 65, 1202–1214. https://doi.org/10.1136/gutjnl-2015-310798.
- 151.Luo, W.; Xu, Q.; Wang, Q.; Wu, H.; Hua, J. Effect of modulation of PPAR-γ activity on Kupffer cells M1/M2 polarization in the development of non-alcoholic fatty liver disease. Sci. Rep. 2017, 7, 44612. https://doi.org/10.1038/srep44612.
- 152.Ahmadian, M.; Suh, J.M.; Hah, N.; Liddle, C.; Atkins, A.R.; Downes, M.; Evans, R.M. PPARγ signaling and metabolism: The good, the bad and the future. Nat. Med. 2013, 19, 557–566. https://doi.org/10.1038/nm.3159.
- 153.Dormandy, J.A.; Charbonnel, B.; Eckland, D.J.; Erdmann, E.; Massi-Benedetti, M.; Moules, I.K.; Skene, A.M.; Tan, M.H.; Lefèbvre, P.J.; Murray, G.D.; et al. Secondary prevention of macrovascular events in patients with type 2 diabetes in the PROactive Study (PROspective pioglitAzone Clinical Trial In macroVascular Events): A randomised controlled trial. Lancet 2005, 366, 1279–1289. https://doi.org/10.1016/s0140-6736(05)67528-9.
- 154.Kirchgessner, T.G.; Sleph, P.; Ostrowski, J.; Lupisella, J.; Ryan, C.S.; Liu, X.; Fernando, G.; Grimm, D.; Shipkova, P.; Zhang, R.; et al. Beneficial and Adverse Effects of an LXR Agonist on Human Lipid and Lipoprotein Metabolism and Circulating Neutrophils. Cell Metab. 2016, 24, 223–233. https://doi.org/10.1016/j.cmet.2016.07.016.
- 155.Muse, E.D.; Yu, S.; Edillor, C.R.; Tao, J.; Spann, N.J.; Troutman, T.D.; Seidman, J.S.; Henke, A.; Roland, J.T.; Ozeki, K.A.; et al. Cell-specific discrimination of desmosterol and desmosterol mimetics confers selective regulation of LXR and SREBP in macrophages. Proc. Natl. Acad. Sci. USA 2018, 115, E4680-e4689. https://doi.org/10.1073/pnas.1714518115.
- 156.EASL-EASD-EASO Clinical Practice Guidelines on the management of metabolic dysfunction-associated steatotic liver disease (MASLD). J. Hepatol. 2024, 81, 492–542. https://doi.org/10.1016/j.jhep.2024.04.031.
How to Cite
Sui, X.; Wu, M.; Yang, X.; Wang, Y. The Multifaced Roles of Hepatic Macrophages in MASLD. Health and Metabolism 2024, 1 (1), 4. https://doi.org/10.53941/hm.2024.100004.
RIS
BibTex
Copyright & License

Copyright (c) 2024 by the authors.
This work is licensed under a Creative Commons Attribution 4.0 International License.
Contents
References


