CRISPR-Cas system is an adaptive immune system of prokaryotes against foreign invading viruses. Type II CRISPR-Cas9 system, as the earliest of the Class II systems to be discovered and applied to gene editing, has been recently found to possess trans-cleavage activity with other Class II systems not long ago. In this review, we summarize the molecular mechanism of cis- and trans-cleavage of target nucleic acids by the Class II type II CRISPR-Cas system, introduce the nucleic acid detection platforms developed based on its trans-cleavage activity, and compare them with those developed based on the type V, type VI, and type I CRISPR-Cas systems.
- Open Access
- Review
Cas9 Cis/Trans Cleavage Mechanisms: Cross-System CRISPR Insights and Diagnostic Applications
- Xueying Li,
- Ying Chen,
- Linglong Huang,
- Jiyun Chen *,
- Liang Liu *
Author Information
Received: 17 Mar 2025 | Revised: 11 Apr 2025 | Accepted: 15 May 2025 | Published: 16 Jul 2025
Abstract
Graphical Abstract
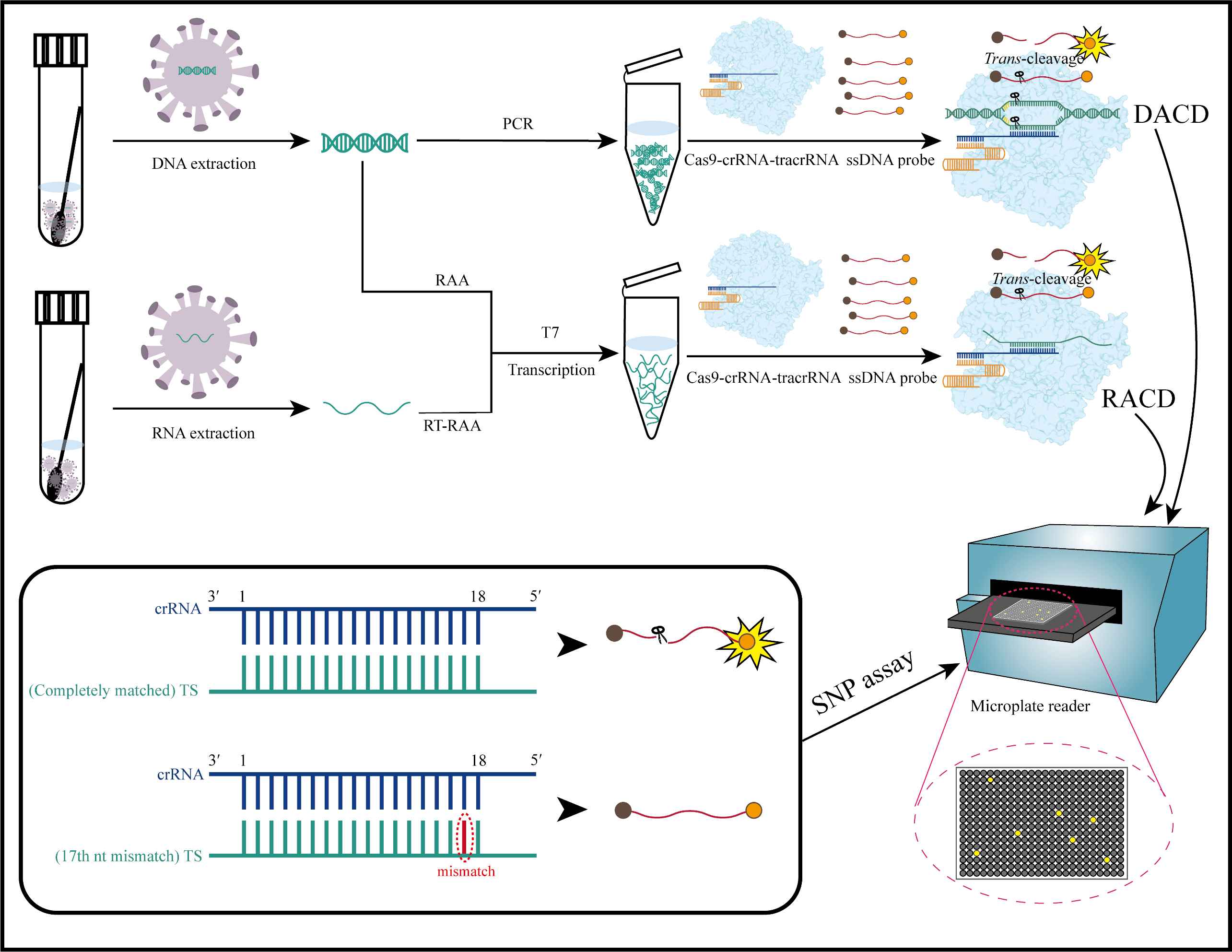
Keywords
CRISPR-Cas | Cas9 | cis-cleavage | trans-cleavage | diagnostics
References
- 1.Ishino, Y.; Shinagawa, H.; Makino, K.; Amemura, M.; Nakata, A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 1987, 169, 5429–5433. https://doi.org/10.1128/jb.169.12.5429-5433.1987.
- 2.Jansen, R.; Embden, J.D.; Gaastra, W.; Schouls, L.M. Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 2002, 43, 1565–1575. https://doi.org/10.1046/j.1365-2958.2002.02839.x.
- 3.Mojica, F.J.; Díez-Villaseñor, C.; García-Martínez, J.; Soria, E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 2005, 60, 174–182. https://doi.org/10.1007/s00239-004-0046-3.
- 4.Bolotin, A.; Quinquis, B.; Sorokin, A.; Ehrlich, S.D. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 2005, 151, 2551–2561. https://doi.org/10.1099/mic.0.28048-0.
- 5.Pourcel, C.; Salvignol, G.; Vergnaud, G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 2005, 151, 653–663. https://doi.org/10.1099/mic.0.27437-0.
- 6.Mojica, F.J.M.; Díez-Villaseñor, C.; García-Martínez, J.; Almendros, C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 2009, 155, 733–740. https://doi.org/10.1099/mic.0.023960-0.
- 7.Deltcheva, E.; Chylinski, K.; Sharma, C.M.; Gonzales, K.; Chao, Y.; Pirzada, Z.A.; Eckert, M.R.; Vogel, J.; Charpentier, E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 2011, 471, 602. https://doi.org/10.1038/nature09886.
- 8.Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. https://doi.org/10.1126/science.1225829.
- 9.Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-guided human genome engineering via Cas9. Science 2013, 339, 823–826. https://doi.org/10.1126/science.1232033.
- 10.Cong, L.; Ran, F.A.; Cox, D.; Lin, S.L.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.B.; Jiang, W.Y.; Marraffini, L.A.; et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 2013, 339, 819–823. https://doi.org/10.1126/science.1231143.
- 11.Cho, S.W.; Kim, S.; Kim, J.M.; Kim, J.S. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013, 31, 230–232. https://doi.org/10.1038/nbt.2507.
- 12.Jinek, M.; East, A.; Cheng, A.; Lin, S.; Ma, E.; Doudna, J. RNA-programmed genome editing in human cells. Elife 2013, 2, e00471. https://doi.org/10.7554/eLife.00471.
- 13.Horvath, P.; Barrangou, R. CRISPR/Cas, the Immune System of Bacteria and Archaea. Science 2010, 327, 167–170. https://doi.org/10.1126/science.1179555.
- 14.Makarova, K.S.; Haft, D.H.; Barrangou, R.; Brouns, S.J.; Charpentier, E.; Horvath, P.; Moineau, S.; Mojica, F.J.; Wolf, Y.I.; Yakunin, A.F.; et al. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 2011, 9, 467–477. https://doi.org/10.1038/nrmicro2577.
- 15.Chylinski, K.; Le Rhun, A.; Charpentier, E. The tracrRNA and Cas9 families of type II CRISPR-Cas immunity systems. RNA Biol. 2013, 10, 726–737. https://doi.org/10.4161/rna.24321.
- 16.Fonfara, I.; Le Rhun, A.; Chylinski, K.; Makarova, K.S.; Lécrivain, A.-L.; Bzdrenga, J.; Koonin, E.V.; Charpentier, E. Phylogeny of Cas9 determines functional exchangeability of dual-RNA and Cas9 among orthologous type II CRISPR-Cas systems. Nucleic Acids Res. 2013, 42, 2577–2590. https://doi.org/10.1093/nar/gkt1074.
- 17.Altae-Tran, H.; Kannan, S.; Demircioglu, F.E.; Oshiro, R.; Nety, S.P.; McKay, L.J.; Dlakić, M.; Inskeep, W.P.; Makarova, K.S.; Macrae, R.K.; et al. The widespread IS200/IS605 transposon family encodes diverse programmable RNA-guided endonucleases. Science 2021, 374, 57–65. https://doi.org/10.1126/science.abj6856.
- 18.Makarova, K.S.; Wolf, Y.I.; Iranzo, J.; Shmakov, S.A.; Alkhnbashi, O.S.; Brouns, S.J.J.; Charpentier, E.; Cheng, D.; Haft, D.H.; Horvath, P.; et al. Evolutionary classification of CRISPR-Cas systems: A burst of class 2 and derived variants. Nat. Rev. Microbiol. 2020, 18, 67–83. https://doi.org/10.1038/s41579-019-0299-x.
- 19.Jiang, F.G.; Doudna, J.A. CRISPR-Cas9 Structures and Mechanisms. Annu. Rev. Biophys. 2017, 46, 505–529.
- 20.Goltsman, D.S.A.; Alexander, L.M.; Lin, J.-L.; Fregoso Ocampo, R.; Freeman, B.; Lamothe, R.C.; Perez Rivas, A.; Temoche-Diaz, M.M.; Chadha, S.; Nordenfelt, N.; et al. Compact Cas9d and HEARO enzymes for genome editing discovered from uncultivated microbes. Nat. Commun. 2022, 13, 7602. https://doi.org/10.1038/s41467-022-35257-7.
- 21.Wang, J.Y.; Pausch, P.; Doudna, J.A. Structural biology of CRISPR-Cas immunity and genome editing enzymes. Nat. Rev. Microbiol. 2022, 20, 641–656. https://doi.org/10.1038/s41579-022-00739-4.
- 22.Jinek, M.; Jiang, F.; Taylor, D.W.; Sternberg, S.H.; Kaya, E.; Ma, E.; Anders, C.; Hauer, M.; Zhou, K.; Lin, S.; et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science 2014, 343, 1247997. https://doi.org/10.1126/science.1247997.
- 23.Nishimasu, H.; Cong, L.; Yan, W.X.; Ran, F.A.; Zetsche, B.; Li, Y.; Kurabayashi, A.; Ishitani, R.; Zhang, F.; Nureki, O. Crystal Structure of Staphylococcus aureus Cas9. Cell 2015, 162, 1113–1126. https://doi.org/10.1016/j.cell.2015.08.007.
- 24.Zhang, S.; Zhang, Q.; Hou, X.M.; Guo, L.; Wang, F.; Bi, L.; Zhang, X.; Li, H.H.; Wen, F.; Xi, X.G.; et al. Dynamics of Staphylococcus aureus Cas9 in DNA target Association and Dissociation. EMBO Rep. 2020, 21, e50184. https://doi.org/10.15252/embr.202050184.
- 25.Du, W.; Zhu, H.; Qian, J.; Xue, D.; Zheng, S.; Huang, Q. Full-Length Model of SaCas9-sgRNA-DNA Complex in Cleavage State. Int. J. Mol. Sci. 2023, 24, 1204. https://doi.org/10.3390/ijms24021204.
- 26.Hirano, H.; Gootenberg, J.S.; Horii, T.; Abudayyeh, O.O.; Kimura, M.; Hsu, P.D.; Nakane, T.; Ishitani, R.; Hatada, I.; Zhang, F.; et al. Structure and Engineering of Francisella novicida Cas9. Cell 2016, 164, 950–961. https://doi.org/10.1016/j.cell.2016.01.039.
- 27.Acharya, S.; Ansari, A.H.; Kumar Das, P.; Hirano, S.; Aich, M.; Rauthan, R.; Mahato, S.; Maddileti, S.; Sarkar, S.; Kumar, M.; et al. PAM-flexible Engineered FnCas9 variants for robust and ultra-precise genome editing and diagnostics. Nat. Commun. 2024, 15, 5471. https://doi.org/10.1038/s41467-024-49233-w.
- 28.Acharya, S.; Mishra, A.; Paul, D.; Ansari, A.H.; Azhar, M.; Kumar, M.; Rauthan, R.; Sharma, N.; Aich, M.; Sinha, D.; et al. Francisella novicida Cas9 interrogates genomic DNA with very high specificity and can be used for mammalian genome editing. Proc. Natl. Acad. Sci. USA 2019, 116, 20959–20968. https://doi.org/10.1073/pnas.1818461116.
- 29.Edraki, A.; Mir, A.; Ibraheim, R.; Gainetdinov, I.; Yoon, Y.; Song, C.-Q.; Cao, Y.; Gallant, J.; Xue, W.; Rivera-Perez, J.A.; et al. A Compact, High-Accuracy Cas9 with a Dinucleotide PAM for In vivo Genome Editing. Mol. Cell 2019, 73, 714. https://doi.org/10.1016/j.molcel.2018.12.003.
- 30.Kim, E.; Koo, T.; Park, S.W.; Kim, D.; Kim, K.; Cho, H.-Y.; Song, D.W.; Lee, K.J.; Jung, M.H.; Kim, S.; et al. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat. Commun. 2017, 8, 14500. https://doi.org/10.1038/ncomms14500.
- 31.Ocampo, R.F.; Bravo, J.P.K.; Dangerfield, T.L.; Nocedal, I.; Jirde, S.A.; Alexander, L.M.; Thomas, N.C.; Das, A.; Nielson, S.; Johnson, K.A.; et al. DNA targeting by compact Cas9d and its resurrected ancestor. Nat. Commun. 2025, 16, 457. https://doi.org/10.1038/s41467-024-55573-4.
- 32.Paul, B.; Montoya, G. CRISPR-Cas12a: Functional overview and applications. Biomed. J. 2020, 43, 8–17. https://doi.org/10.1016/j.bj.2019.10.005.
- 33.Zetsche, B.; Gootenberg, J.S.; Abudayyeh, O.O.; Slaymaker, I.M.; Makarova, K.S.; Essletzbichler, P.; Volz, S.E.; Joung, J.; van der Oost, J.; Regev, A.; et al. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell 2015, 163, 759–771. https://doi.org/10.1016/j.cell.2015.09.038.
- 34.East-Seletsky, A.; O’Connell, M.R.; Knight, S.C.; Burstein, D.; Cate, J.H.D.; Tjian, R.; Doudna, J.A. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature 2016, 538, 270. https://doi.org/10.1038/nature19802.
- 35.Abudayyeh, O.O.; Gootenberg, J.S.; Essletzbichler, P.; Han, S.; Joung, J.; Belanto, J.J.; Verdine, V.; Cox, D.B.T.; Kellner, M.J.; Regev, A.; et al. RNA targeting with CRISPR-Cas13. Nature 2017, 550, 280. https://doi.org/10.1038/nature24049.
- 36.Hu, C.; Ni, D.; Nam, K.H.; Majumdar, S.; McLean, J.; Stahlberg, H.; Terns, M.P.; Ke, A. Allosteric control of type I-A CRISPR-Cas3 complexes and establishment as effective nucleic acid detection and human genome editing tools. Mol. Cell 2022, 82, 2754. https://doi.org/10.1016/j.molcel.2022.06.007.
- 37.Guo, J.; Gong, L.; Yu, H.; Li, M.; An, Q.; Liu, Z.; Fan, S.; Yang, C.; Zhao, D.; Han, J.; et al. Engineered minimal type I CRISPR-Cas system for transcriptional activation and base editing in human cells. Nat. Commun. 2024, 15, 7277. https://doi.org/10.1038/s41467-024-51695-x.
- 38.Anders, C.; Niewoehner, O.; Duerst, A.; Jinek, M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 2014, 513, 569. https://doi.org/10.1038/nature13579.
- 39.Yamano, T.; Nishimasu, H.; Zetsche, B.; Hirano, H.; Slaymaker, I.M.; Li, Y.; Fedorova, I.; Nakane, T.; Makarova, K.S.; Koonin, E.V.; et al. Crystal Structure of Cpf1 in Complex with Guide RNA and Target DNA. Cell 2016, 165, 949–962. https://doi.org/10.1016/j.cell.2016.04.003.
- 40.Babu, K.; Amrani, N.; Jiang, W.; Yogesha, S.D.; Nguyen, R.; Qin, P.Z.; Rajan, R. Bridge Helix of Cas9 Modulates Target DNA Cleavage and Mismatch Tolerance. Biochemistry 2019, 58, 1905–1917. https://doi.org/10.1021/acs.biochem.8b01241.
- 41.Gasiunas, G.; Barrangou, R.; Horvath, P.; Siksnys, V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. USA 2012, 109, E2579–E2586. https://doi.org/10.1073/pnas.1208507109.
- 42.Zhu, X.; Clarke, R.; Puppala, A.K.; Chittori, S.; Merk, A.; Merrill, B.J.; Simonovic, M.; Subramaniam, S. Cryo-EM structures reveal coordinated domain motions that govern DNA cleavage by Cas9. Nat. Struct. Mol. Biol. 2019, 26, 679. https://doi.org/10.1038/s41594-019-0258-2.
- 43.Jiang, F.G.; Zhou, K.H.; Ma, L.L.; Gressel, S.; Doudna, J.A. A Cas9-guide RNA complex preorganized for target DNA recognition. Science 2015, 348, 1477–1481. https://doi.org/10.1126/science.aab1452.
- 44.Hsu, P.D.; Scott, D.A.; Weinstein, J.A.; Ran, F.A.; Konermann, S.; Agarwala, V.; Li, Y.; Fine, E.J.; Wu, X.; Shalem, O.; et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013, 31, 827. https://doi.org/10.1038/nbt.2647.
- 45.Sternberg, S.H.; Redding, S.; Jinek, M.; Greene, E.C.; Doudna, J.A. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 2014, 507, 62. https://doi.org/10.1038/nature13011.
- 46.Jiang, F.; Taylor, D.W.; Chen, J.S.; Kornfeld, J.E.; Zhou, K.; Thompson, A.J.; Nogales, E.; Doudna, J.A. Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science 2016, 351, 867–871. https://doi.org/10.1126/science.aad8282.
- 47.Gong, S.; Yu, H.H.; Johnson, K.A.; Taylor, D.W. DNA Unwinding Is the Primary Determinant of CRISPR-Cas9 Activity. Cell Rep. 2018, 22, 359–371. https://doi.org/10.1016/j.celrep.2017.12.041.
- 48.Zeng, Y.; Cui, Y.; Zhang, Y.; Zhang, Y.; Liang, M.; Chen, H.; Lan, J.; Song, G.; Lou, J. The initiation, propagation and dynamics of CRISPR-SpyCas9 R-loop complex. Nucleic Acids Res. 2018, 46, 350–361. https://doi.org/10.1093/nar/gkx1117.
- 49.Yang, M.; Peng, S.; Sun, R.; Lin, J.; Wang, N.; Chen, C. The Conformational Dynamics of Cas9 Governing DNA Cleavage Are Revealed by Single-Molecule FRET. Cell Rep. 2018, 22, 372–382. https://doi.org/10.1016/j.celrep.2017.12.048.
- 50.Pacesa, M.; Loeff, L.; Querques, I.; Muckenfuss, L.M.; Sawicka, M.; Jinek, M. R-loop formation and conformational activation mechanisms of Cas9. Nature 2022, 609, 191. https://doi.org/10.1038/s41586-022-05114-0.
- 51.Szczelkun, M.D.; Tikhomirova, M.S.; Sinkunas, T.; Gasiunas, G.; Karvelis, T.; Pschera, P.; Siksnys, V.; Seidel, R. Direct observation of R-loop formation by single RNA-guided Cas9 and Cascade effector complexes. Proc. Natl. Acad. Sci. USA 2014, 111, 9798–9803. https://doi.org/10.1073/pnas.1402597111.
- 52.Wu, X.; Kriz, A.J.; Sharp, P.A. Target specificity of the CRISPR-Cas9 system. Quant. Biol. 2014, 2, 59–70. https://doi.org/10.1007/s40484-014-0030-x.
- 53.Yu, T.; Liu, T.; Wang, Y.; Zhao, X.; Zhang, W. Effect of Cas9 Protein on the Seed-Target Base Pair of the sgRNA/DNA Hybrid Duplex. J. Phys. Chem. B 2023, 127, 4989–4997. https://doi.org/10.1021/acs.jpcb.3c00997.
- 54.Sternberg, S.H.; LaFrance, B.; Kaplan, M.; Doucina, J.A. Conformational control of DNA target cleavage by CRISPR-Cas9. Nature 2015, 527, 110–113. https://doi.org/10.1038/nature15544.
- 55.Babu, K.; Kathiresan, V.; Kumari, P.; Newsom, S.; Parameshwaran, H.P.; Chen, X.; Liu, J.; Qin, P.Z.; Rajan, R. Coordinated Actions of Cas9 HNH and RuvC Nuclease Domains Are Regulated by the Bridge Helix and the Target DNA Sequence. Biochemistry 2021, 60, 3783–3800. https://doi.org/10.1021/acs.biochem.1c00354.
- 56.Nierzwicki, L.; East, K.W.; Binz, J.M.; Hsu, R.V.; Ahsan, M.; Arantes, P.R.; Skeens, E.; Pacesa, M.; Jinek, M.; Lisi, G.P.; et al. Principles of target DNA cleavage and the role of Mg2+ in the catalysis of CRISPR-Cas9. Nat. Catal. 2022, 5, 912–922. https://doi.org/10.1038/s41929-022-00848-6.
- 57.Nishimasu, H.; Ran, F.A.; Hsu, P.D.; Konermann, S.; Shehata, S.I.; Dohmae, N.; Ishitani, R.; Zhang, F.; Nureki, O. Crystal Structure of Cas9 in Complex with Guide RNA and Target DNA. Cell 2014, 156, 935–949. https://doi.org/10.1016/j.cell.2014.02.001.
- 58.Casalino, L.; Nierzwicki, L.; Jinek, M.; Palermo, G. Catalytic Mechanism of Non-Target DNA Cleavage in CRISPR-Cas9 Revealed by Ab Initio Molecular Dynamics. Acs Catal. 2020, 10, 13596–13605. https://doi.org/10.1021/acscatal.0c03566.
- 59.Shou, J.; Li, J.; Liu, Y.; Wu, Q. Precise and Predictable CRISPR Chromosomal Rearrangements Reveal Principles of Cas9-Mediated Nucleotide Insertion. Mol. Cell 2018, 71, 498–509.e494. https://doi.org/10.1016/j.molcel.2018.06.021.
- 60.Shi, X.; Shou, J.; Mehryar, M.M.; Li, J.; Wang, L.; Zhang, M.; Huang, H.; Sun, X.; Wu, Q. Cas9 has no exonuclease activity resulting in staggered cleavage with overhangs and predictable di- and tri-nucleotide CRISPR insertions without template donor. Cell Discov. 2019, 5, 53. https://doi.org/10.1038/s41421-019-0120-z.
- 61.Zuo, Z.; Liu, J. Cas9-catalyzed DNA Cleavage Generates Staggered Ends: Evidence from Molecular Dynamics Simulations. Sci. Rep. 2016, 5, 37584. https://doi.org/10.1038/srep37584.
- 62.Chen, J.; Chen, Y.; Huang, L.; Lin, X.; Chen, H.; Xiang, W.; Liu, L. Trans-nuclease activity of Cas9 activated by DNA or RNA target binding. Nat. Biotechnol. 2024, 43, 558–568. https://doi.org/10.1038/s41587-024-02255-7.
- 63.Strohkendl, I.; Saha, A.; Moy, C.; Nguyen, A.H.; Ahsan, M.; Russell, R.; Palermo, G.; Taylor, D.W. Cas12a domain flexibility guides R-loop formation and forces RuvC resetting. Mol. Cell 2024, 84, 2717–2731.e6. https://doi.org/10.1016/j.molcel.2024.06.007.
- 64.Liu, L.; Li, X.Y.; Wang, J.Y.; Wang, M.; Chen, P.; Yin, M.L.; Li, J.Z.; Sheng, G.; Wang, Y.L. Two Distant Catalytic Sites Are Responsible for C2c2 RNase Activities. Cell 2017, 168, 121. https://doi.org/10.1016/j.cell.2016.12.031.
- 65.Adler, B.A.; Trinidad, M.I.; Bellieny-Rabelo, D.; Zhang, E.; Karp, H.M.; Skopintsev, P.; Thornton, B.W.; Weissman, R.F.; Yoon, P.H.; Chen, L.; et al. CasPEDIA Database: A functional classification system for class 2 CRISPR-Cas enzymes. Nucleic Acids Res. 2023, Early Access. https://doi.org/10.1093/nar/gkad890.
- 66.Tran, H.A.; Shmakov, S.A.; Makarova, K.S.; Wolf, Y.I.; Kannan, S.; Zhang, F.; Koonin, E.V. Diversity, evolution, and classification of the RNA- guided nucleases TnpB and Cas12. Proc. Natl. Acad. Sci. USA 2023, 120, e2308224120. https://doi.org/10.1073/pnas.2308224120.
- 67.Duan, Z.; Zhang, X.; Zhang, J.T.; Ji, X.; Liu, R.; Chen, Y.; Li, S.; Jia, N.; Gao, H.; Xin, Y.; et al. Structure and genome editing activity of the novel CRISPR-Cas12o1 effector. Cell Res. 2025, 35, 145–148. https://doi.org/10.1038/s41422-024-01050-y.
- 68.Yan, W.X.; Hunnewell, P.; Alfonse, L.E.; Carte, J.M.; Keston-Smith, E.; Sothiselvam, S.; Garrity, A.J.; Chong, S.R.; Makarova, K.S.; Koonin, E.V.; et al. Functionally diverse type V CRISPR-Cas systems. Science 2019, 363, 88. https://doi.org/10.1126/science.aav7271.
- 69.Wu, W.Y.; Adiego-Perez, B.; van der Oost, J. Biology and applications of CRISPR-Cas12 and transposon-associated homologs. Nat. Biotechnol. 2024, 42, 1807–1821. https://doi.org/10.1038/s41587-024-02485-9.
- 70.Nakagawa, R.; Hirano, H.; Omura, S.N.; Nety, S.; Kannan, S.; Altae-Tran, H.; Yao, X.; Sakaguchi, Y.; Ohira, T.; Wu, W.Y.; et al. Cryo-EM structure of the transposon-associated TnpB enzyme. Nature 2023, 616, 390. https://doi.org/10.1038/s41586-023-05933-9.
- 71.Kato, K.; Okazaki, S.; Kannan, S.; Altae-Tran, H.; Demircioglu, F.E.; Isayama, Y.; Ishikawa, J.; Fukuda, M.; Macrae, R.K.; Nishizawa, T.; et al. Structure of the IscB-ωRNA ribonucleoprotein complex, the likely ancestor of CRISPR-Cas9. Nat. Commun. 2022, 13, 6719. https://doi.org/10.1038/s41467-022-34378-3.
- 72.Swarts, D.C.; van der Oost, J.; Jinek, M. Structural Basis for Guide RNA Processing and Seed-Dependent DNA Targeting by CRISPR-Cas12a. Mol. Cell 2017, 66, 221. https://doi.org/10.1016/j.molcel.2017.03.016.
- 73.Bravo, J.P.K.; Hallmark, T.; Naegle, B.; Beisel, C.L.; Jackson, R.N.; Taylor, D.W. RNA targeting unleashes indiscriminate nuclease activity of CRISPR-Cas12a2. Nature 2023, 613, 582–587. https://doi.org/10.1038/s41586-022-05560-w.
- 74.Dmytrenko, O.; Neumann, G.C.; Hallmark, T.; Keiser, D.J.; Crowley, V.M.; Vialetto, E.; Mougiakos, I.; Wandera, K.G.; Domgaard, H.; Weber, J.; et al. Cas12a2 elicits abortive infection through RNA-triggered destruction of dsDNA. Nature 2023, 613, 588. https://doi.org/10.1038/s41586-022-05559-3.
- 75.Chen, J.S.; Ma, E.; Harrington, L.B.; Da Costa, M.; Tian, X.; Palefsky, J.M.; Doudna, J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 2018, 360, 436. https://doi.org/10.1126/science.aar6245.
- 76.Swarts, D.C.; Jinek, M. Mechanistic Insights into the cis- and trans-Acting DNase Activities of Cas12a. Mol. Cell 2019, 73, 589. https://doi.org/10.1016/j.molcel.2018.11.021.
- 77.Kurihara, N.; Nakagawa, R.; Hirano, H.; Okazaki, S.; Tomita, A.; Kobayashi, K.; Kusakizako, T.; Nishizawa, T.; Yamashita, K.; Scott, D.A.; et al. Structure of the type V-C CRISPR-Cas effector enzyme. Mol. Cell 2022, 82, 1865. https://doi.org/10.1016/j.molcel.2022.03.006.
- 78.Urbaitis, T.; Gasiunas, G.; Young, J.K.; Hou, Z.; Paulraj, S.; Godliauskaite, E.; Juskeviciene, M.M.; Stitilyte, M.; Jasnauskaite, M.; Mabuchi, M.; et al. A new family of CRISPR-type V nucleases with C-rich PAM recognition. Embo Rep. 2022, 23, e55481. https://doi.org/10.15252/embr.202255481.
- 79.Yang, H.; Patel, D.J. Structures, mechanisms and applications of RNA-centric CRISPR-Cas13. Nat. Chem. Biol. 2024, 20, 673–688. https://doi.org/10.1038/s41589-024-01593-6.
- 80.Abudayyeh, O.O.; Gootenberg, J.S.; Konermann, S.; Joung, J.; Slaymaker, I.M.; Cox, D.B.; Shmakov, S.; Makarova, K.S.; Semenova, E.; Minakhin, L.; et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 2016, 353, aaf5573. https://doi.org/10.1126/science.aaf5573.
- 81.Shmakov, S.; Abudayyeh, O.O.; Makarova, K.S.; Wolf, Y.I.; Gootenberg, J.S.; Semenova, E.; Minakhin, L.; Joung, J.; Konermann, S.; Severinov, K.; et al. Discovery and Functional Characterization of Diverse Class 2 CRISPR-Cas Systems. Mol. Cell 2015, 60, 385–397. https://doi.org/10.1016/j.molcel.2015.10.008.
- 82.Barrangou, R.; Marraffini, L.A. CRISPR-Cas systems: Prokaryotes upgrade to adaptive immunity. Mol. Cell 2014, 54, 234–244. https://doi.org/10.1016/j.molcel.2014.03.011.
- 83.Koonin, E.V.; Makarova, K.S. Origins and evolution of CRISPR-Cas systems. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2019, 374, 20180087. https://doi.org/10.1098/rstb.2018.0087.
- 84.Xu, Z.; Li, Y.; Li, M.; Xiang, H.; Yan, A. Harnessing the type I CRISPR-Cas systems for genome editing in prokaryotes. Environ. Microbiol. 2021, 23, 542–558. https://doi.org/10.1111/1462-2920.15116.
- 85.Yoshimi, K.; Takeshita, K.; Kodera, N.; Shibumura, S.; Yamauchi, Y.; Omatsu, M.; Umeda, K.; Kunihiro, Y.; Yamamoto, M.; Mashimo, T. Dynamic mechanisms of CRISPR interference by Escherichia coli CRISPR-Cas3. Nat. Commun. 2022, 13, 4917. https://doi.org/10.1038/s41467-022-32618-0.
- 86.Yoshimi, K.; Takeshita, K.; Yamayoshi, S.; Shibumura, S.; Yamauchi, Y.; Yamamoto, M.; Yotsuyanagi, H.; Kawaoka, Y.; Mashimo, T. CRISPR-Cas3-based diagnostics for SARS-CoV-2 and influenza virus. Iscience 2022, 25, 103830. https://doi.org/10.1016/j.isci.2022.103830.
- 87.He, L.; St John James, M.; Radovcic, M.; Ivancic-Bace, I.; Bolt, E.L. Cas3 Protein-A Review of a Multi-Tasking Machine. Genes. 2020, 11, 208. https://doi.org/10.3390/genes11020208.
- 88.Liu, T.Y.; Doudna, J.A. Chemistry of Class 1 CRISPR-Cas effectors: Binding, editing, and regulation. J. Biol. Chem. 2020, 295, 14473–14487. https://doi.org/10.1074/jbc.REV120.007034.
- 89.Shangguan, Q.; Graham, S.; Sundaramoorthy, R.; White, M.F. Structure and mechanism of the type I-G CRISPR effector. Nucleic Acids Res. 2022, 50, 11214–11228. https://doi.org/10.1093/nar/gkac925.
- 90.Yang, Z.; Li, Z.; Li, B.; Bu, R.; Tan, G.-Y.; Wang, Z.; Yan, H.; Xin, Z.; Zhang, G.; Li, M.; et al. A thermostable type I-B CRISPR-Cas system for orthogonal and multiplexed genetic engineering. Nat. Commun. 2023, 14, 6193. https://doi.org/10.1038/s41467-023-41973-5.
- 91.Govindarajan, S.; Borges, A.; Karambelkar, S.; Bondy-Denomy, J. Distinct Subcellular Localization of a Type I CRISPR Complex and the Cas3 Nuclease in Bacteria. J. Bacteriol. 2022, 204, e00105-22. https://doi.org/10.1128/jb.00105-22.
- 92.Rollins, M.F.; Chowdhury, S.; Carter, J.; Golden, S.M.; Miettinen, H.M.; Santiago-Frangos, A.; Faith, D.; Lawrence, C.M.; Lander, G.C.; Wiedenheft, B. Structure Reveals a Mechanism of CRISPR-RNA-Guided Nuclease Recruitment and Anti-CRISPR Viral Mimicry. Mol. Cell 2019, 74, 132. https://doi.org/10.1016/j.molcel.2019.02.001.
- 93.Gootenberg, J.S.; Abudayyeh, O.O.; Lee, J.W.; Essletzbichler, P.; Dy, A.J.; Joung, J.; Verdine, V.; Donghia, N.; Daringer, N.M.; Freije, C.A.; et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 2017, 356, 438. https://doi.org/10.1126/science.aam9321.
- 94.Li, S.Y.; Cheng, Q.X.; Wang, J.M.; Li, X.Y.; Zhang, Z.L.; Gao, S.; Cao, R.B.; Zhao, G.P.; Wang, J. CRISPR-Cas12a-assisted nucleic acid detection. Cell Discov. 2018, 4, 20. https://doi.org/10.1038/s41421-018-0028-z.
- 95.Harrington, L.B.; Burstein, D.; Chen, J.S.; Paez-Espino, D.; Ma, E.; Witte, I.P.; Cofsky, J.C.; Kyrpides, N.C.; Banfield, J.F.; Doudna, J.A. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science 2018, 362, 839. https://doi.org/10.1126/science.aav4294.
- 96.Gootenberg, J.S.; Abudayyeh, O.O.; Kellner, M.J.; Joung, J.; Collins, J.J.; Zhang, F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 2018, 360, 439. https://doi.org/10.1126/science.aaq0179.
- 97.Teng, F.; Guo, L.; Cui, T.T.; Wang, X.G.; Xu, K.; Gao, Q.Q.; Zhou, Q.; Li, W. CDetection: CRISPR-Cas12b-based DNA detection with sub-attomolar sensitivity and single-base specificity. Genome Biol. 2019, 20, 132. https://doi.org/10.1186/s13059-019-1742-z.
- 98.Li, L.; Li, S.; Wu, N.; Wu, J.; Wang, G.; Zhao, G.; Wang, J. HOLMESv2: A CRISPR-Cas12b-Assisted Platform for Nucleic Acid Detection and DNA Methylation Quantitation. Acs Synth. Biol. 2019, 8, 2228–2237. https://doi.org/10.1021/acssynbio.9b00209.
- 99.Wang, B.; Wang, R.; Wang, D.; Wu, J.; Li, J.; Wang, J.; Liu, H.; Wang, Y. Cas12aVDet: A CRISPR/Cas12a-Based Platform for Rapid and Visual Nucleic Acid Detection. Anal. Chem. 2019, 91, 12156–12161. https://doi.org/10.1021/acs.analchem.9b01526.
- 100.Hu, T.; Ji, Q.; Ke, X.; Zhou, H.; Zhang, S.; Ma, S.; Yu, C.; Ju, W.; Lu, M.; Lin, Y.; et al. Repurposing Type I-A CRISPR-Cas3 for a robust diagnosis of human papillomavirus (HPV). Commun. Biol. 2024, 7, 858. https://doi.org/10.1038/s42003-024-06537-3.
- 101.Wang, R.; Qian, C.; Pang, Y.; Li, M.; Yang, Y.; Ma, H.; Zhao, M.; Qian, F.; Yu, H.; Liu, Z.; et al. opvCRISPR: One-pot visual RT-LAMP-CRISPR platform for SARS-cov-2 detection. Biosens. Bioelectron. 2021, 172, 112766. https://doi.org/10.1016/j.bios.2020.112766.
- 102.Nguyen, L.T.; Gurijala, J.; Rananaware, S.R.; Pizzano, B.L.M.; Stone, B.T.; Jain, P.K. CRISPR-ENHANCE: An enhanced nucleic acid detection platform using Cas12a. Methods 2022, 203, 116–124. https://doi.org/10.1016/j.ymeth.2021.02.001.
- 103.Zeng, Q.; Zhou, M.; Hu, Z.; Deng, W.; Li, Z.; Wu, L.; Liang, D. Rapid and sensitive Cas12a-based one-step nucleic acid detection with ssDNA-modified crRNA. Anal. Chim. Acta 2023, 1276, 341622. https://doi.org/10.1016/j.aca.2023.341622.
- 104.Zeng, Q.L.; Zhou, M.J.; Deng, W.H.; Gao, Q.; Li, Z.; Wu, L.Q.; Liang, D.S. Sensitive and visual detection of SARS-CoV-2 using RPA-Cas12a one-step assay with ssDNA-modified crRNA. Anal. Chim. Acta 2024, 1309, 342693. https://doi.org/10.1016/j.aca.2024.342693.
- 105.Hu, M.; Qiu, Z.; Bi, Z.; Tian, T.; Jiang, Y.; Zhou, X. Photocontrolled crRNA activation enables robust CRISPR-Cas12a diagnostics. Proc. Natl. Acad. Sci. USA 2022, 119, e2202034119. https://doi.org/10.1073/pnas.2202034119.
- 106.Hu, M.; Liu, R.; Qiu, Z.; Cao, F.; Tian, T.; Lu, Y.; Jiang, Y.; Zhou, X. Light-Start CRISPR-Cas12a Reaction with Caged crRNA Enables Rapid and Sensitive Nucleic Acid Detection. Angew. Chem. Int. Ed. 2023, 62, e202300663. https://doi.org/10.1002/anie.202300663.
- 107.Ackerman, C.M.; Myhrvold, C.; Thakku, S.G.; Freije, C.A.; Metsky, H.C.; Yang, D.K.; Ye, S.H.; Boehm, C.K.; Kosoko-Thoroddsen, T.-S. F.; et al. Massively multiplexed nucleic acid detection with Cas13. Nature 2020, 582, 277. https://doi.org/10.1038/s41586-020-2279-8.
- 108.van Dongen, J.E.; Berendsen, J.T.W.; Steenbergen, R.D.M.; Wolthuis, R.M.F.; Eijkel, J.C.T.; Segerink, L.I. Point-of-care CRISPR/Cas nucleic acid detection: Recent advances, challenges and opportunities. Biosens. Bioelectron. 2020, 166. https://doi.org/10.1016/j.bios.2020.112445.
- 109.Weng, Z.; You, Z.; Yang, J.; Mohammad, N.; Lin, M.; Wei, Q.; Gao, X.; Zhang, Y. CRISPR-Cas Biochemistry and CRISPR-Based Molecular Diagnostics. Angew. Chem. Int. Ed. 2023, 62, e202214987. https://doi.org/10.1002/anie.202214987.
- 110.Li, H.M.; Xie, Y.; Chen, F.M.; Bai, H.W.; Xiu, L.S.; Zhou, X.N.; Guo, X.K.; Hu, Q.Q.; Yin, K. Amplification-free CRISPR/Cas detection technology: Challenges, strategies, and perspectives. Chem. Soc. Rev. 2023, 52, 361–382. https://doi.org/10.1039/d2cs00594h.
- 111.Iida, T.; Shinoda, H.; Watanabe, R. SATORI: Amplification-free digital RNA detection method for the diagnosis of viral infections. Biophys. Physicobiol. 2023, 20, e200031. https://doi.org/10.2142/biophysico.bppb-v20.0031.
- 112.Yin, L.; Man, S.; Ye, S.; Liu, G.; Ma, L. CRISPR-Cas based virus detection: Recent advances and perspectives. Biosens. Bioelectron. 2021, 193, 113541. https://doi.org/10.1016/j.bios.2021.113541.
- 113.Feng, W.; Newbigging, A.M.; Tao, J.; Cao, Y.; Peng, H.; Le, C.; Wu, J.; Pang, B.; Li, J.; Tyrrell, D.L.; et al. CRISPR technology incorporating amplification strategies: Molecular assays for nucleic acids, proteins, and small molecules. Chem. Sci. 2021, 12, 4683–4698. https://doi.org/10.1039/d0sc06973f.
- 114.Iwasaki, R.S.; Batey, R.T. SPRINT: A Cas13a-based platform for detection of small molecules. Nucleic Acids Res. 2020, 48, e101. https://doi.org/10.1093/nar/gkaa673.
- 115.Sun, Y.; Yu, L.; Liu, C.; Ye, S.; Chen, W.; Li, D.; Huang, W. One-tube SARS-CoV-2 detection platform based on RT-RPA and CRISPR/Cas12a. J. Transl. Med. 2021, 19, 74. https://doi.org/10.1186/s12967-021-02741-5.
- 116.Sheng, Y.; Zhang, T.; Zhang, S.; Johnston, M.; Zheng, X.; Shan, Y.; Liu, T.; Huang, Z.; Qian, F.; Xie, Z.; et al. A CRISPR/Cas13a-powered catalytic electrochemical biosensor for successive and highly sensitive RNA diagnostics. Biosens. Bioelectron. 2021, 178, 113027. https://doi.org/10.1016/j.bios.2021.113027.
- 117.Liu, T.Y.; Knott, G.J.; Smock, D.C.J.; Desmarais, J.J.; Son, S.; Bhuiya, A.; Jakhanwal, S.; Prywes, N.; Agrawal, S.; Derby, M.D.D.; et al. Accelerated RNA detection using tandem CRISPR nucleases. Nat. Chem. Biol. 2021, 17, 982–988. https://doi.org/10.1038/s41589-021-00842-2.
- 118.Tang, Y.; Qi, L.; Liu, Y.; Guo, L.; Zhao, R.; Yang, M.; Du, Y.; Li, B. CLIPON: A CRISPR-Enabled Strategy that Turns Commercial Pregnancy Test Strips into General Point-of-Need Test Devices. Angew. Chem. Int. Ed. 2022, 134, e202115907. https://doi.org/10.1002/anie.202115907.
- 119.Broto, M.; Kaminski, M.M.; Adrianus, C.; Kim, N.; Greensmith, R.; Dissanayake-Perera, S.; Schubert, A.J.; Tan, X.; Kim, H.; Dighe, A.S.; et al. Nanozyme-catalysed CRISPR assay for preamplification-free detection of non-coding RNAs. Nat. Nanotechnol. 2022, 17, 1120–1126. https://doi.org/10.1038/s41565-022-01179-0.
- 120.Yang, J.; Song, Y.; Deng, X.; Vanegas, J.A.; You, Z.; Zhang, Y.; Weng, Z.; Avery, L.; Dieckhaus, K.D.; Peddi, A.; et al. Engineered LwaCas13a with enhanced collateral activity for nucleic acid detection. Nat. Chem. Biol. 2023, 19, 45–54. https://doi.org/10.1038/s41589-022-01135-y.
- 121.Chen, J.; Lin, X.; Xiang, W.; Chen, Y.; Zhao, Y.; Huang, L.; Liu, L. DNA target binding-induced pre-crRNA processing in type II and V CRISPR-Cas systems. Nucleic Acids Res. 2025, 53, gkae1241. https://doi.org/10.1093/nar/gkae1241.
- 122.Dai, J.; Wu, B.; Ai, F.; Yang, Z.; Lu, Y.; Zinian, C.; Zeng, K.; Zhang, Z. Exploiting the Potential of Spherical PAM Antenna for Enhanced CRISPR-Cas12a: A Paradigm Shift toward a Universal Amplification-Free Nucleic Acid Test Platform. Anal. Chem. 2025, 97, 1236–1245. https://doi.org/10.1021/acs.analchem.4c04871.
- 123.Richardson, C.D.; Ray, G.J.; DeWitt, M.A.; Curie, G.L.; Corn, J.E. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat. Biotechnol. 2016, 34, 339–344. https://doi.org/10.1038/nbt.3481.
- 124.Feng, Y.; Liu, S.; Chen, R.; Xie, A. Target binding and residence: A new determinant of DNA double-strand break repair pathway choice in CRISPR/Cas9 genome editing. J. Zhejiang Univ. Sci. B 2021, 22, 73–86. https://doi.org/10.1631/jzus.B2000282.
How to Cite
Li, X.; Chen, Y.; Huang, L.; Chen, J.; Liu, L. Cas9 Cis/Trans Cleavage Mechanisms: Cross-System CRISPR Insights and Diagnostic Applications. Health and Metabolism 2025, 2 (3), 6. https://doi.org/10.53941/hm.2025.100021.
RIS
BibTex
Copyright & License

Copyright (c) 2025 by the authors.
This work is licensed under a Creative Commons Attribution 4.0 International License.
Contents
References


