Riboswitches are natural RNA regulatory elements that control gene expression through ligand-induced conformational changes. These dynamic RNA sensors modulate downstream gene activity by influencing transcription or translation. Recent advances in riboswitch engineering have expanded their utility in biotechnology and synthetic biology. This review highlights key insights into the structural and functional dynamics of riboswitches and discusses emerging strategies for their application as programmable biological tools.
- Open Access
- Review
Riboswitch Dynamics and Their Expanding Biotechnological Applications
- Dian Chen,
- Yu Liu *
Author Information
Received: 06 Jun 2025 | Revised: 27 Jun 2025 | Accepted: 23 Jul 2025 | Published: 13 Aug 2025
Abstract
Graphical Abstract
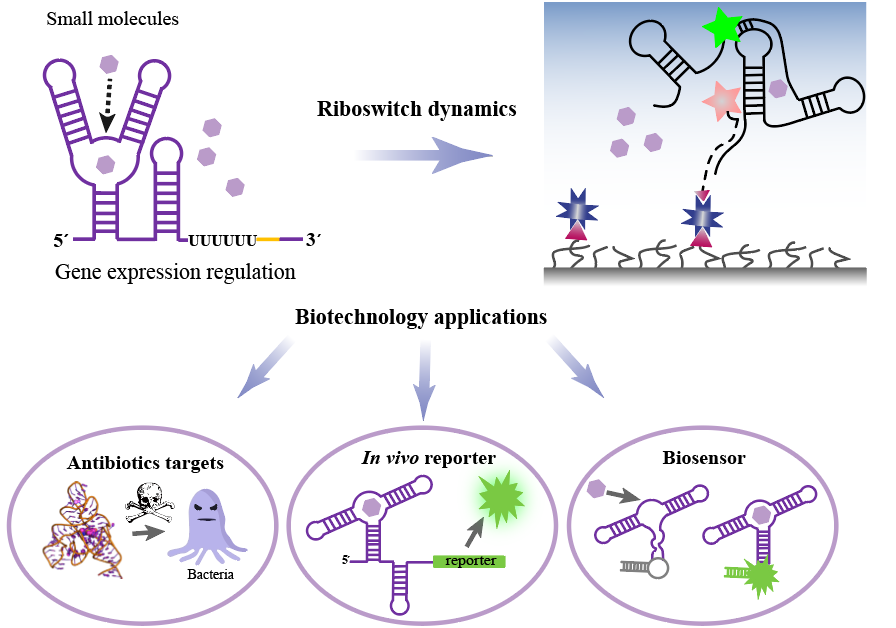
Keywords
RNA | riboswitch | dynamics | applications
References
- 1.Liang, X.; Chen, D.; Su, A.; Liu, Y. Divergent Molecular Assembly and Catalytic Mechanisms between Bacterial and Archaeal RNase P in Pre-tRNA Cleavage. Proc. Natl. Acad. Sci. USA 2024, 121, e2407579121. https://doi.org/10.1073/pnas.2407579121.
- 2.Dar, D.; Sorek, R. Regulation of Antibiotic-Resistance by Non-Coding RNAs in Bacteria. Curr. Opin. Microbiol. 2017, 36, 111–117. https://doi.org/10.1016/j.mib.2017.02.005.
- 3.Liu, B.; Samaniego, C.C.; Bennett, M.R.; Franco, E.; Chappell, J. A Portable Regulatory RNA Array Design Enables Tunable and Complex Regulation across Diverse Bacteria. Nat. Commun. 2023, 14, 5268. https://doi.org/10.1038/s41467-023-40785-x.
- 4.Schmerer, N.; Janga, H.; Aillaud, M.; Hoffmann, J.; Aznaourova, M.; Wende, S.; Steding, H.; Halder, L.D.; Uhl, M.; Boldt, F.; et al. A Searchable Atlas of Pathogen-Sensitive lncRNA Networks in Human Macrophages. Nat. Commun. 2025, 16, 4733. https://doi.org/10.1038/s41467-025-60084-x.
- 5.Wilson, R.C.; Doudna, J.A. Molecular Mechanisms of RNA Interference. Annu. Rev. Biophys. 2013, 42, 217–239. https://doi.org/10.1146/annurev-biophys-083012-130404.
- 6.Nugent, P.J.; Park, H.; Wladyka, C.L.; Yelland, J.N.; Sinha, S.; Chen, K.Y.; Bynum, C.; Quarterman, G.; Lee, S.C.; Hsieh, A.C.; et al. Decoding Post-Transcriptional Regulatory Networks by RNA-Linked CRISPR Screening in Human Cells. Nat. Methods 2025, 22, 1237–1246. https://doi.org/10.1038/s41592-025-02702-6.
- 7.Gao, L.; Chen, D.; Liu, Y. Ligand Response of Guanidine-IV Riboswitch at Single-Molecule Level. eLife 2024, 13, RP94706. https://doi.org/10.7554/eLife.94706.
- 8.Waters, L.S. Bacterial Manganese Sensing and Homeostasis. Curr. Opin. Chem. Biol. 2020, 55, 96–102. https://doi.org/10.1016/j.cbpa.2020.01.003.
- 9.Guanzon, D.A.; Pienkoß, S.; Brandenburg, V.B.; Röder, J.; Scheller, D.; Dietze, A.; Wimbert, A.; Twittenhoff, C.; Narberhaus, F. Two Temperature-Responsive RNAs Act in Concert: The Small RNA CyaR and the mRNA ompX. Nucleic Acids Res. 2025, 53, gkaf041. https://doi.org/10.1093/nar/gkaf041.
- 10.Wu, L.; Chen, D.; Ding, J.; Liu, Y. A Transient Conformation Facilitates Ligand Binding to the Adenine Riboswitch. iScience 2021, 24, 103512. https://doi.org/10.1016/j.isci.2021.103512.
- 11.Atilho, R.M.; Mirihana Arachchilage, G.; Greenlee, E.B.; Knecht, K.M.; Breaker, R.R. A Bacterial Riboswitch Class for the Thiamin Precursor HMP-PP Employs a Terminator-Embedded Aptamer. eLife 2019, 8, e45210. https://doi.org/10.7554/eLife.45210.
- 12.Sudarsan, N.; Lee, E.R.; Weinberg, Z.; Moy, R.H.; Kim, J.N.; Link, K.H.; Breaker, R.R. Riboswitches in Eubacteria Sense the Second Messenger Cyclic Di-GMP. Science 2008, 321, 411–413. https://doi.org/10.1126/science.1159519.
- 13.Zhao, B.; Guffy, S.L.; Williams, B.; Zhang, Q. An Excited State Underlies Gene Regulation of a Transcriptional Riboswitch. Nat. Chem. Biol. 2017, 13, 968–974. https://doi.org/10.1038/nchembio.2427.
- 14.Capdevila, D.A.; Rondón, J.J.; Edmonds, K.A.; Rocchio, J.S.; Dujovne, M.V.; Giedroc, D.P. Bacterial Metallostasis: Metal Sensing, Metalloproteome Remodeling, and Metal Trafficking. Chem. Rev. 2024, 124, 13574–13659. https://doi.org/10.1021/acs.chemrev.4c00264.
- 15.Kiliushik, D.; Goenner, C.; Law, M.; Schroeder, G.M.; Srivastava, Y.; Jenkins, J.L.; Wedekind, J.E. Knotty Is Nice: Metabolite Binding and RNA-Mediated Gene Regulation by the preQ1 Riboswitch Family. J. Biol. Chem. 2024, 300, 107951. https://doi.org/10.1016/j.jbc.2024.107951.
- 16.Chen, D.; Han, Z.; Liang, X.; Liu, Y. Engineering a DNA Polymerase for Modifying Large RNA at Specific Positions. Nat. Chem. 2025, 17, 382–392. https://doi.org/10.1038/s41557-024-01707-6.
- 17.Stagno, J.R.; Wang, Y.-X. Riboswitch Mechanisms for Regulation of P1 Helix Stability. Int. J. Mol. Sci. 2024, 25, 10682. https://doi.org/10.3390/ijms251910682.
- 18.Zheng, L.; Song, Q.; Xu, X.; Shen, X.; Li, C.; Li, H.; Chen, H.; Ren, A. Structure-Based Insights into Recognition and Regulation of SAM-Sensing Riboswitches. Sci. China Life Sci. 2023, 66, 31–50. https://doi.org/10.1007/s11427-022-2188-7.
- 19.Bushhouse, D.Z.; Fu, J.; Lucks, J.B. RNA Folding Kinetics Control Riboswitch Sensitivity in Vivo. Nat. Commun. 2025, 16, 953. https://doi.org/10.1038/s41467-024-55601-3.
- 20.Chen, D.; Li, J.; Wu, Y.; Hong, L.; Liu, Y. Structural Dynamics-Guided Engineering of a Riboswitch RNA for Evolving c-Di-AMP Synthases. Sci. Adv. 2025, 11, eadt8165. https://doi.org/10.1126/sciadv.adt8165.
- 21.Hedwig, V.; Spöring, M.; Ottlinger, J.; Köse, S.; Nar, H.; Schnapp, G.; Gottschling, D.; Klein, H.; Aspnes, G.; Klugmann, M.; et al. Engineering Oxypurinol-Responsive Riboswitches Based on Bacterial Xanthine Aptamers for Gene Expression Control in Mammalian Cell Culture. Nucleic Acids Res. 2025, 53, gkae1189. https://doi.org/10.1093/nar/gkae1189.
- 22.Ogawa, A.; Fujikawa, M.; Tanimoto, R.; Matsuno, K.; Uehara, R.; Inoue, H.; Takahashi, H. Cell-Free Multistep Gene Regulatory Cascades Using Eukaryotic ON-Riboswitches Responsive to in Situ Expressed Protein Ligands. ACS Synth. Biol. 2025, 14, 909–918. https://doi.org/10.1021/acssynbio.4c00840.
- 23.Pham, H.L.; Wong, A.; Chua, N.; Teo, W.S.; Yew, W.S.; Chang, M.W. Engineering a Riboswitch-Based Genetic Platform for the Self-Directed Evolution of Acid-Tolerant Phenotypes. Nat. Commun. 2017, 8, 411. https://doi.org/10.1038/s41467-017-00511-w.
- 24.Serganov, A.; Huang, L.; Patel, D.J. Coenzyme Recognition and Gene Regulation by a Flavin Mononucleotide Riboswitch. Nature 2009, 458, 233–237. https://doi.org/10.1038/nature07642.
- 25.Degenhardt, M.F.S.; Degenhardt, H.F.; Bhandari, Y.R.; Lee, Y.-T.; Ding, J.; Yu, P.; Heinz, W.F.; Stagno, J.R.; Schwieters, C.D.; Watts, N.R.; et al. Determining Structures of RNA Conformers Using AFM and Deep Neural Networks. Nature 2025, 637, 1234–1243. https://doi.org/10.1038/s41586-024-07559-x.
- 26.Haack, D.B.; Rudolfs, B.; Jin, S.; Khitun, A.; Weeks, K.M.; Toor, N. Scaffold-Enabled High-Resolution Cryo-EM Structure Determination of RNA. Nat. Commun. 2025, 16, 880. https://doi.org/10.1038/s41467-024-55699-5.
- 27.Wang, S.; Chen, D.; Gao, L.; Liu, Y. Short Oligonucleotides Facilitate Co-Transcriptional Labeling of RNA at Specific Positions. J. Am. Chem. Soc. 2022, 144, 5494–5502. https://doi.org/10.1021/jacs.2c00020.
- 28.Wickiser, J.K.; Winkler, W.C.; Breaker, R.R.; Crothers, D.M. The Speed of RNA Transcription and Metabolite Binding Kinetics Operate an FMN Riboswitch. Mol. Cell 2005, 18, 49–60. https://doi.org/10.1016/j.molcel.2005.02.032.
- 29.Winkler, W.; Nahvi, A.; Breaker, R.R. Thiamine Derivatives Bind Messenger RNAs Directly to Regulate Bacterial Gene Expression. Nature 2002, 419, 952–956. https://doi.org/10.1038/nature01145.
- 30.Winkler, W.C.; Nahvi, A.; Roth, A.; Collins, J.A.; Breaker, R.R. Control of Gene Expression by a Natural Metabolite-Responsive Ribozyme. Nature 2004, 428, 281–286. https://doi.org/10.1038/nature02362.
- 31.Reining, A.; Nozinovic, S.; Schlepckow, K.; Buhr, F.; Fürtig, B.; Schwalbe, H. Three-State Mechanism Couples Ligand and Temperature Sensing in Riboswitches. Nature 2013, 499, 355–359. https://doi.org/10.1038/nature12378.
- 32.Manz, C.; Kobitski, A.Y.; Samanta, A.; Keller, B.G.; Jäschke, A.; Nienhaus, G.U. Single-Molecule FRET Reveals the Energy Landscape of the Full-Length SAM-I Riboswitch. Nat. Chem. Biol. 2017, 13, 1172–1178. https://doi.org/10.1038/nchembio.2476.
- 33.Neupane, K.; Yu, H.; Foster, D.A.N.; Wang, F.; Woodside, M.T. Single-Molecule Force Spectroscopy of the Add Adenine Riboswitch Relates Folding to Regulatory Mechanism. Nucleic Acids Res. 2011, 39, 7677–7687. https://doi.org/10.1093/nar/gkr305.
- 34.Stagno, J.R.; Liu, Y.; Bhandari, Y.R.; Conrad, C.E.; Panja, S.; Swain, M.; Fan, L.; Nelson, G.; Li, C.; Wendel, D.R.; et al. Structures of Riboswitch RNA Reaction States by Mix-and-Inject XFEL Serial Crystallography. Nature 2017, 541, 242–246. https://doi.org/10.1038/nature20599.
- 35.Zhang, K.; Li, S.; Kappel, K.; Pintilie, G.; Su, Z.; Mou, T.-C.; Schmid, M.F.; Das, R.; Chiu, W. Cryo-EM Structure of a 40 kDa SAM-IV Riboswitch RNA at 3.7 Å Resolution. Nat. Commun. 2019, 10, 5511. https://doi.org/10.1038/s41467-019-13494-7.
- 36.Reyes, F.E.; Schwartz, C.R.; Tainer, J.A.; Rambo, R.P. Methods for Using New Conceptual Tools and Parameters to Assess RNA Structure by Small-Angle X-Ray Scattering. Methods Enzymol. 2014, 549, 235–263. https://doi.org/10.1016/B978-0-12-801122-5.00011-8.
- 37.Baird, N.J.; Ferré-D’Amaré, A.R. Idiosyncratically Tuned Switching Behavior of Riboswitch Aptamer Domains Revealed by Comparative Small-Angle X-Ray Scattering Analysis. RNA 2010, 16, 598–609. https://doi.org/10.1261/rna.1852310.
- 38.Roy, S.; Lammert, H.; Hayes, R.L.; Chen, B.; LeBlanc, R.; Dayie, T.K.; Onuchic, J.N.; Sanbonmatsu, K.Y. A Magnesium-Induced Triplex Pre-Organizes the SAM-II Riboswitch. PLoS Comput. Biol. 2017, 13, e1005406. https://doi.org/10.1371/journal.pcbi.1005406.
- 39.Kulshina, N.; Baird, N.J.; Ferré-D’Amaré, A.R. Recognition of the Bacterial Second Messenger Cyclic Diguanylate by Its Cognate Riboswitch. Nat. Struct. Mol. Biol. 2009, 16, 1212–1217. https://doi.org/10.1038/nsmb.1701.
- 40.Crielaard, S.; Peters, C.F.M.; Slivkov, A.; van den Homberg, D.A.L.; Velema, W.A. Chemotranscriptomic Profiling with a Thiamine Monophosphate Photoaffinity Probe. Chem. Sci. 2025, 16, 4725–4731. https://doi.org/10.1039/d4sc06189f.
- 41.Eschbach, S.H.; Hien, E.D.M.; Ghosh, T.; Lamontagne, A.-M.; Lafontaine, D.A. The Escherichia Coli ribB Riboswitch Senses Flavin Mononucleotide within a Defined Transcriptional Window. RNA 2024, 30, 1660–1673. https://doi.org/10.1261/rna.080074.124.
- 42.Ott, E.; Stolz, J.; Lehmann, M.; Mack, M. The RFN Riboswitch of Bacillus Subtilis Is a Target for the Antibiotic Roseoflavin Produced by Streptomyces Davawensis. RNA Biol. 2009, 6, 276–280. https://doi.org/10.4161/rna.6.3.8342.
- 43.Mansjö, M.; Johansson, J. The Riboflavin Analog Roseoflavin Targets an FMN-Riboswitch and Blocks Listeria Monocytogenes Growth, but Also Stimulates Virulence Gene-Expression and Infection. RNA Biol. 2011, 8, 674–680. https://doi.org/10.4161/rna.8.4.15586.
- 44.Howe, J.A.; Wang, H.; Fischmann, T.O.; Balibar, C.J.; Xiao, L.; Galgoci, A.M.; Malinverni, J.C.; Mayhood, T.; Villafania, A.; Nahvi, A.; et al. Selective Small-Molecule Inhibition of an RNA Structural Element. Nature 2015, 526, 672–677. https://doi.org/10.1038/nature15542.
- 45.Rizvi, N.F.; Howe, J.A.; Nahvi, A.; Klein, D.J.; Fischmann, T.O.; Kim, H.-Y.; McCoy, M.A.; Walker, S.S.; Hruza, A.; Richards, M.P.; et al. Discovery of Selective RNA-Binding Small Molecules by Affinity-Selection Mass Spectrometry. ACS Chem. Biol. 2018, 13, 820–831. https://doi.org/10.1021/acschembio.7b01013.
- 46.Kim, J.N.; Blount, K.F.; Puskarz, I.; Lim, J.; Link, K.H.; Breaker, R.R. Design and Antimicrobial Action of Purine Analogues That Bind Guanine Riboswitches. ACS Chem. Biol. 2009, 4, 915–927. https://doi.org/10.1021/cb900146k.
- 47.Mulhbacher, J.; Brouillette, E.; Allard, M.; Fortier, L.-C.; Malouin, F.; Lafontaine, D.A. Novel Riboswitch Ligand Analogs as Selective Inhibitors of Guanine-Related Metabolic Pathways. PLoS Pathog. 2010, 6, e1000865. https://doi.org/10.1371/journal.ppat.1000865.
- 48.Gilbert, S.D.; Mediatore, S.J.; Batey, R.T. Modified Pyrimidines Specifically Bind the Purine Riboswitch. J. Am. Chem. Soc. 2006, 128, 14214–14215. https://doi.org/10.1021/ja063645t.
- 49.Kim, J.N.; Blount, K.F.; Puskarz, I.; Lim, J.; Link, K.H.; Breaker, R.R. Design and Antimicrobial Action of Purine Analogues That Bind Guanine Riboswitches. ACS Chem. Biol. 2009, 4, 915–927. https://doi.org/10.1021/cb900146k.
- 50.Childs-Disney, J.L.; Yang, X.; Gibaut, Q.M.R.; Tong, Y.; Batey, R.T.; Disney, M.D. Targeting RNA Structures with Small Molecules. Nat. Rev. Drug Discov. 2022, 21, 736–762. https://doi.org/10.1038/s41573-022-00521-4.
- 51.Falese, J.P.; Donlic, A.; Hargrove, A.E. Targeting RNA with Small Molecules: From Fundamental Principles towards the Clinic. Chem. Soc. Rev. 2021, 50, 2224–2243. https://doi.org/10.1039/d0cs01261k.
- 52.Gutierrez-Preciado, A.; Jensen, R.A.; Yanofsky, C.; Merino, E. New Insights into Regulation of the Tryptophan Biosynthetic Operon in Gram-Positive Bacteria. Trends Genet. TIG 2005, 21, 432–436. https://doi.org/10.1016/j.tig.2005.06.001.
- 53.Campos-Chavez, E.; Paul, S.; Zhou, Z.; Alonso, D.; Verma, A.R.; Fei, J.; Mondragón, A. Translational T-Box Riboswitches Bind tRNA by Modulating Conformational Flexibility. Nat. Commun. 2024, 15, 6592. https://doi.org/10.1038/s41467-024-50885-x.
- 54.Suddala, K.C.; Yoo, J.; Fan, L.; Zuo, X.; Wang, Y.-X.; Chung, H.S.; Zhang, J. Direct Observation of tRNA-Chaperoned Folding of a Dynamic mRNA Ensemble. Nat. Commun. 2023, 14, 5438. https://doi.org/10.1038/s41467-023-41155-3.
- 55.Niu, X.; Xu, Z.; Zhang, Y.; Zuo, X.; Chen, C.; Fang, X. Structural and Dynamic Mechanisms for Coupled Folding and tRNA Recognition of a Translational T-Box Riboswitch. Nat. Commun. 2023, 14, 7394. https://doi.org/10.1038/s41467-023-43232-z.
- 56.Orac, C.M.; Zhou, S.; Means, J.A.; Boehm, D.; Bergmeier, S.C.; Hines, J.V. Synthesis and Stereospecificity of 4,5-Disubstituted Oxazolidinone Ligands Binding to T-Box Riboswitch RNA. J. Med. Chem. 2011, 54, 6786–6795. https://doi.org/10.1021/jm2006904.
- 57.Anupam, R.; Denapoli, L.; Muchenditsi, A.; Hines, J.V. Identification of Neomycin B-Binding Site in T Box Antiterminator Model RNA. Bioorg. Med. Chem. 2008, 16, 4466–4470. https://doi.org/10.1016/j.bmc.2008.02.056.
- 58.Fowler, C.C.; Brown, E.D.; Li, Y. Using a Riboswitch Sensor to Examine Coenzyme B(12) Metabolism and Transport in E. Coli. Chem. Biol. 2010, 17, 756–765. https://doi.org/10.1016/j.chembiol.2010.05.025.
- 59.Xue, Y.; Li, J.; Chen, D.; Zhao, X.; Hong, L.; Liu, Y. Observation of Structural Switch in Nascent SAM-VI Riboswitch during Transcription at Single-Nucleotide and Single-Molecule Resolution. Nat. Commun. 2023, 14, 2320. https://doi.org/10.1038/s41467-023-38042-2.
- 60.Kipkorir, T.; Polgar, P.; Barker, D.; D’Halluin, A.; Patel, Z.; Arnvig, K.B. A Novel Regulatory Interplay between Atypical B12 Riboswitches and uORF Translation in Mycobacterium Tuberculosis. Nucleic Acids Res. 2024, 52, 7876–7892. https://doi.org/10.1093/nar/gkae338.
- 61.Gao, X.; Dong, X.; Subramanian, S.; Matthews, P.M.; Cooper, C.A.; Kearns, D.B.; Dann, C.E. Engineering of Bacillus Subtilis Strains to Allow Rapid Characterization of Heterologous Diguanylate Cyclases and Phosphodiesterases. Appl. Environ. Microbiol. 2014, 80, 6167–6174. https://doi.org/10.1128/AEM.01638-14.
- 62.Michener, J.K.; Smolke, C.D. High-Throughput Enzyme Evolution in Saccharomyces Cerevisiae Using a Synthetic RNA Switch. Metab. Eng. 2012, 14, 306–316. https://doi.org/10.1016/j.ymben.2012.04.004.
- 63.Wang, J.; Gao, D.; Yu, X.; Li, W.; Qi, Q. Evolution of a Chimeric Aspartate Kinase for L-Lysine Production Using a Synthetic RNA Device. Appl. Microbiol. Biotechnol. 2015, 99, 8527–8536. https://doi.org/10.1007/s00253-015-6615-0.
- 64.Meyer, A.; Pellaux, R.; Potot, S.; Becker, K.; Hohmann, H.-P.; Panke, S.; Held, M. Optimization of a Whole-Cell Biocatalyst by Employing Genetically Encoded Product Sensors inside Nanolitre Reactors. Nat. Chem. 2015, 7, 673–678. https://doi.org/10.1038/nchem.2301.
- 65.Truong, L.; Ferré-D’Amaré, A.R. From Fluorescent Proteins to Fluorogenic RNAs: Tools for Imaging Cellular Macromolecules. Protein Sci. Publ. Protein Soc. 2019, 28, 1374–1386. https://doi.org/10.1002/pro.3632.
- 66.You, M.; Litke, J.L.; Jaffrey, S.R. Imaging Metabolite Dynamics in Living Cells Using a Spinach-Based Riboswitch. Proc. Natl. Acad. Sci. USA 2015, 112, E2756–E2765. https://doi.org/10.1073/pnas.1504354112.
- 67.Paige, J.S.; Nguyen-Duc, T.; Song, W.; Jaffrey, S.R. Fluorescence Imaging of Cellular Metabolites with RNA. Science 2012, 335, 1194. https://doi.org/10.1126/science.1218298.
- 68.Bose, D.; Su, Y.; Marcus, A.; Raulet, D.H.; Hammond, M.C. An RNA-Based Fluorescent Biosensor for High-Throughput Analysis of the cGAS-cGAMP-STING Pathway. Cell Chem. Biol. 2016, 23, 1539–1549. https://doi.org/10.1016/j.chembiol.2016.10.014.
- 69.Chen, Z.; Chen, W.; Reheman, Z.; Jiang, H.; Wu, J.; Li, X. Genetically Encoded RNA-Based Sensors with Pepper Fluorogenic Aptamer. Nucleic Acids Res. 2023, 51, 8322–8336. https://doi.org/10.1093/nar/gkad620.
- 70.Kellenberger, C.A.; Chen, C.; Whiteley, A.T.; Portnoy, D.A.; Hammond, M.C. RNA-Based Fluorescent Biosensors for Live Cell Imaging of Second Messenger Cyclic Di-AMP. J. Am. Chem. Soc. 2015, 137, 6432–6435. https://doi.org/10.1021/jacs.5b00275.
- 71.Wang, X.C.; Wilson, S.C.; Hammond, M.C. Next-Generation RNA-Based Fluorescent Biosensors Enable Anaerobic Detection of Cyclic Di-GMP. Nucleic Acids Res. 2016, 44, e139. https://doi.org/10.1093/nar/gkw580.
- 72.Braselmann, E.; Palmer, A.E. A Multicolor Riboswitch-Based Platform for Imaging of RNA in Live Mammalian Cells. Methods Enzymol. 2020, 641, 343–372. https://doi.org/10.1016/bs.mie.2020.03.004.
- 73.Braselmann, E.; Wierzba, A.J.; Polaski, J.T.; Chromiński, M.; Holmes, Z.E.; Hung, S.-T.; Batan, D.; Wheeler, J.R.; Parker, R.; Jimenez, R.; et al. A Multicolor Riboswitch-Based Platform for Imaging of RNA in Live Mammalian Cells. Nat. Chem. Biol. 2018, 14, 964–971. https://doi.org/10.1038/s41589-018-0103-7.
- 74.Bühler, B.; Schokolowski, J.; Benderoth, A.; Englert, D.; Grün, F.; Jäschke, A.; Sunbul, M. Avidity-Based Bright and Photostable Light-up Aptamers for Single-Molecule mRNA Imaging. Nat. Chem. Biol. 2023, 19, 478–487. https://doi.org/10.1038/s41589-022-01228-8.
- 75.Baker, J.L.; Sudarsan, N.; Weinberg, Z.; Roth, A.; Stockbridge, R.B.; Breaker, R.R. Widespread Genetic Switches and Toxicity Resistance Proteins for Fluoride. Science 2012, 335, 233–235. https://doi.org/10.1126/science.1215063.
- 76.Thavarajah, W.; Silverman, A.D.; Verosloff, M.S.; Kelley-Loughnane, N.; Jewett, M.C.; Lucks, J.B. Point-of-Use Detection of Environmental Fluoride via a Cell-Free Riboswitch-Based Biosensor. ACS Synth. Biol. 2020, 9, 10–18. https://doi.org/10.1021/acssynbio.9b00347.
- 77.Ariyarathna, M.R.; Nissanka, J.P.; Methlal, K.; Abeyrathne, K.D.; Satharasinghe, M.; Banushan, P.; Manawadu, D.; Mirihana Arachchilage, G.; Silva, G.N. Simple and Cost-Effective Fluoride Riboswitch-Based Whole-Cell Biosensor for the Determination of Fluoride in Drinking Water. Appl. Biochem. Biotechnol. 2025. https://doi.org/10.1007/s12010-025-05269-2.
- 78.Brown, D.M.; Phillips, D.A.; Garcia, D.C.; Arce, A.; Lucci, T.; Davies, J.P.; Mangini, J.T.; Rhea, K.A.; Bernhards, C.B.; Biondo, J.R.; et al. Semiautomated Production of Cell-Free Biosensors. ACS Synth. Biol. 2025, 14, 979–986. https://doi.org/10.1021/acssynbio.4c00703.
- 79.Boyd, M.A.; Thavarajah, W.; Lucks, J.B.; Kamat, N.P. Robust and Tunable Performance of a Cell-Free Biosensor Encapsulated in Lipid Vesicles. Sci. Adv. 2023, 9, eadd6605. https://doi.org/10.1126/sciadv.add6605.
- 80.Galizi, R.; Jaramillo, A. Engineering CRISPR Guide RNA Riboswitches for in Vivo Applications. Curr. Opin. Biotechnol. 2019, 55, 103–113. https://doi.org/10.1016/j.copbio.2018.08.007.
- 81.Hu, L.-F.; Li, Y.-X.; Wang, J.-Z.; Zhao, Y.-T.; Wang, Y. Controlling CRISPR-Cas9 by Guide RNA Engineering. Wiley Interdiscip. Rev. RNA 2023, 14, e1731. https://doi.org/10.1002/wrna.1731.
- 82.Fukunaga, K.; Teramoto, T.; Nakashima, M.; Ohtani, T.; Katsuki, R.; Matsuura, T.; Yokobayashi, Y.; Kakuta, Y. Structural Insights into Lab-Coevolved RNA-RBP Pairs and Applications of Synthetic Riboswitches in Cell-Free System. Nucleic Acids Res. 2025, 53, gkaf212. https://doi.org/10.1093/nar/gkaf212.
How to Cite
Chen, D.; Liu, Y. Riboswitch Dynamics and Their Expanding Biotechnological Applications. Health and Metabolism 2025, 2 (4), 4. https://doi.org/10.53941/hm.2025.100027.
RIS
BibTex
Copyright & License

Copyright (c) 2025 by the authors.
This work is licensed under a Creative Commons Attribution 4.0 International License.
Contents
References


