As the fundamental unit of life, the metabolic state of an individual cell is regarded as a direct indicator of its physiological and pathological processes. Live single-cell mass spectrometry (Live-SCMS) has emerged as a key technique at the intersection of metabolomics and single-cell biology, with recent advancements in sampling strategies, signal detection, and biocompatibility. Conventional single-cell metabolite detection methods based on bulk-cell or fixed-cell analyses are often destructive and preclude further cellular analysis. In contrast, Live-SCMS enables in situ and dynamic profiling of cellular metabolism while preserving cellular integrity and viability. This review systematically outlines the core strategies of Live-SCMS, including minimally invasive sampling, low-perturbation workflows in buffered environments, biocompatible labeling, and isotope tracing. It emphasizes the unique advantages of the technique over traditional methods, such as higher sensitivity, improved spatial resolution, and real-time analysis capability, all achieved without compromising cell viability. Recent applications in cancer metabolism, drug response, immune cell functional profiling, and studies of plants, embryos, and microbial models are summarized. Future perspectives on technological advancement, broader biological applications, and clinical relevance are also provided
- Open Access
- Review
Enabling Live Single-Cell Metabolomics: Mass Spectrometry Technologies and Functional Applications
- Simin Cheng 1,2,
- Danni Wu 2,
- Xiaoxiao Ma 2,*,
- Xiaoyun Gong 1,*
Author Information
Received: 23 Jul 2025 | Revised: 06 Sep 2025 | Accepted: 18 Sep 2025 | Published: 12 Jan 2026
Abstract
Graphical Abstract
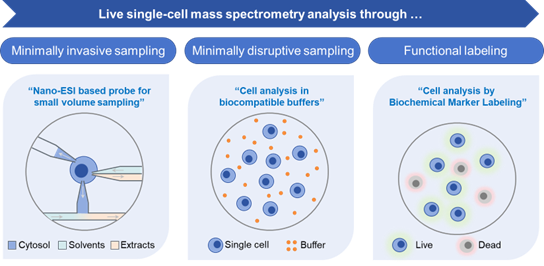
Keywords
References
- 1.
Zenobi, R. Single-Cell Metabolomics: Analytical and Biological Perspectives. Science 2013, 342, 1243259. https://doi.org/10.1126/science.1243259.
- 2.
Steven, J.A.; Lani, F.W. Cellular Heterogeneity: Do Differences Make a Difference? Cell 2010, 141, 559–563. https://doi.org/10.1016/j.cell.2010.04.033.
- 3.
Kinker, G.S.; Greenwald, A.C.; Tal, R.; Orlova, Z.; Cuoco, M.S.; McFarland, J.M.; Warren, A.; Rodman, C.; Roth, J.A.; Bender, S.A.; et al. Pan-Cancer Single-Cell RNA-Seq Identifies Recurring Programs of Cellular Heterogeneity. Nat. Genet. 2020, 52, 1208–1218. https://doi.org/10.1038/s41588-020-00726-6.
- 4.
Kitano, H. Systems Biology: A Brief Overview. Science 2002, 295, 1662–1664. https://doi.org/10.1126/science.1069492.
- 5.
Marshall, J.L.; Peshkin, B.N.; Yoshino, T.; Vowinckel, J.; Danielsen, H.E.; Melino, G.; Tsamardinos, I.; Haudenschild, C.; Kerr, D.J.; Sampaio, C.; et al. The Essentials of Multiomics. Oncologist 2022, 27, 272–284. https://doi.org/10.1093/oncolo/oyab048.
- 6.
Wu, R.Q.; Zhao, X.F.; Wang, Z.Y.; Zhou, M.; Chen, Q.M. Novel Molecular Events in Oral Carcinogenesis via Integrative Approaches. J. Dent. Res. 2011, 90, 561–572. https://doi.org/10.1177/0022034510383691.
- 7.
Weibel, K.E.; Mor, J.R.; Fiechter, A. Rapid Sampling of Yeast Cells and Automated Assays of Adenylate, Citrate, Pyruvate and Glucose-6-Phosphate Pools. Anal. Biochem. 1974, 58, 208–216. https://doi.org/10.1016/0003-2697(74)90459-x.
- 8.
Taylor, M.J.; Lukowski, J.K.; Anderton, C.R. Spatially Resolved Mass Spectrometry at the Single Cell: Recent Innovations in Proteomics and Metabolomics. J. Am. Soc. Mass Spectrom. 2021, 32, 872–894. https://doi.org/10.1021/jasms.0c00439.
- 9.
Zhao, P.; Feng, Y.; Wu, J.; Zhu, J.; Yang, J.; Ma, X.; Ouyang, Z.; Zhang, X.; Zhang, W.; Wang, W. Efficient Sample Preparation System for Multi-Omics Analysis via Single Cell Mass Spectrometry. Anal. Chem. 2023, 95, 7212–7219. https://doi.org/10.1021/acs.analchem.2c05728.
- 10.
Rubakhin, S.S.; Romanova, E.V.; Nemes, P.; Sweedler, J.V. Profiling Metabolites and Peptides in Single Cells. Nat. Methods 2011, 8, 20–29. https://doi.org/10.1038/nmeth.1549.
- 11.
Heinemann, M.; Zenobi, R. Single Cell Metabolomics. Curr. Opin. Biotechnol. 2011, 22, 26–31. https://doi.org/10.1016/j.copbio.2010.09.008.
- 12.
Shimizu, M.; Levi-Schaffer, F.; Ojima, N.; Shingaki, T.; Masujima, T. A Single-Cell Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass-Spectroscopic Assay of the Cell-Maturation Process. Anal. Sci. 2002, 18, 107–108. https://doi.org/10.2116/analsci.18.107.
- 13.
Xu, S.; Yang, C.; Yan, X.; Liu, H. Towards High Throughput and High Information Coverage: Advanced Single-Cell Mass Spectrometric Techniques. Anal. Bioanal. Chem. 2022, 414, 219–233. https://doi.org/10.1007/s00216-021-03624-w.
- 14.
Hu, R.; Li, Y.; Yang, Y.; Liu, M. Mass Spectrometry‐based Strategies for Single‐cell Metabolomics. Mass Spectrom. Rev. 2023, 42, 67–94. https://doi.org/10.1002/mas.21704.
- 15.
Zhang, L.; Vertes, A. Single‐cell Mass Spectrometry Approaches to Explore Cellular Heterogeneity. Angew. Chem. Int. Ed. 2018, 57, 4466–4477. https://doi.org/10.1002/anie.201709719.
- 16.
Okumoto, S. Imaging Approach for Monitoring Cellular Metabolites and Ions Using Genetically Encoded Biosensors. Curr. Opin. Biotechnol. 2010, 21, 45–54. https://doi.org/10.1016/j.copbio.2010.01.009.
- 17.
Mizuno, H.; Tsuyama, N.; Harada, T.; Masujima, T. Live Single-Cell Video-Mass Spectrometry for Cellular and Subcellular Molecular Detection and Cell Classification. J. Mass Spectrom. 2008, 43, 1692–1700. https://doi.org/10.1002/jms.1460.
- 18.
Tsuyama, N.; Mizuno, H.; Tokunaga, E.; Masujima, T. Live Single-Cell Molecular Analysis by Video-Mass Spectrometry. Anal. Sci. Int. J. JPN Soc. Anal. Chem. 2008, 24, 559–561. https://doi.org/10.2116/analsci.24.559.
- 19.
Fujii, T.; Matsuda, S.; Tejedor, M.L.; Esaki, T.; Sakane, I.; Mizuno, H.; Tsuyama, N.; Masujima, T. Direct Metabolomics for Plant Cells by Live Single-Cell Mass Spectrometry. Nat. Protoc. 2015, 10, 1445–1456. https://doi.org/10.1038/nprot.2015.084.
- 20.
Ali, A.; Abouleila, Y.; Amer, S.; Furushima, R.; Emara, S.; Equis, S.; Cotte, Y.; Masujima, T. Quantitative Live Single-Cell Mass Spectrometry with Spatial Evaluation by Three-Dimensional Holographic and Tomographic Laser Microscopy. Anal. Sci. 2016, 32, 125–127. https://doi.org/10.2116/analsci.32.125.
- 21.
Zhang, L.; Vertes, A. Energy Charge, Redox State, and Metabolite Turnover in Single Human Hepatocytes Revealed by Capillary Microsampling Mass Spectrometry. Anal. Chem. 2015, 87, 10397–10405. https://doi.org/10.1021/acs.analchem.5b02502.
- 22.
Yin, R.; Prabhakaran, V.; Laskin, J. Quantitative Extraction and Mass Spectrometry Analysis at a Single-Cell Level. Anal. Chem. 2018, 90, 7937–7945. https://doi.org/10.1021/acs.analchem.8b00551.
- 23.
Pan, N.; Rao, W.; Kothapalli, N.R.; Liu, R.; Burgett, A.W.G.; Yang, Z. The Single-Probe: A Miniaturized Multifunctional Device for Single Cell Mass Spectrometry Analysis. Anal. Chem. 2014, 86, 9376–9380. https://doi.org/10.1021/ac5029038.
- 24.
Standke, S.J.; Colby, D.H.; Bensen, R.C.; Burgett, A.W.G.; Yang, Z. Mass Spectrometry Measurement of Single Suspended Cells Using a Combined Cell Manipulation System and a Single-Probe Device. Anal. Chem. 2019, 91, 1738–1742. https://doi.org/10.1021/acs.analchem.8b05774.
- 25.
Gong, X.; Zhao, Y.; Cai, S.; Fu, S.; Yang, C.; Zhang, S.; Zhang, X. Single Cell Analysis with Probe ESI-Mass Spectrometry: Detection of Metabolites at Cellular and Subcellular Levels. Anal. Chem. 2014, 86, 3809–3816. https://doi.org/10.1021/ac500882e.
- 26.
Aerts, J.T.; Louis, K.R.; Crandall, S.R.; Govindaiah, G.; Cox, C.L.; Sweedler, J.V. Patch Clamp Electrophysiology and Capillary Electrophoresis-Mass Spectrometry Metabolomics for Single Cell Characterization. Anal. Chem. 2014, 86, 3203–3208. https://doi.org/10.1021/ac500168d.
- 27.
Zhu, H.; Li, Q.; Liao, T.; Yin, X.; Chen, Q.; Wang, Z.; Dai, M.; Yi, L.; Ge, S.; Miao, C.; et al. Metabolomic Profiling of Single Enlarged Lysosomes. Nat. Methods 2021, 18, 788–798. https://doi.org/10.1038/s41592-021-01182-8.
- 28.
Zhang, Q.; Lin, L.; Yi, X.; Xie, T.; Xing, G.; Li, Y.; Wang, X.; Lin, J.-M. Microfluidic Sampling of Undissolved Components from Subcellular Regions of Living Single Cells for Mass Spectrometry Analysis. Anal. Chem. 2023, 95, 18082–18090. https://doi.org/10.1021/acs.analchem.3c03086.
- 29.
Guillaume-Gentil, O.; Rey, T.; Kiefer, P.; Ibáñez, A.J.; Steinhoff, R.; Brönnimann, R.; Dorwling-Carter, L.; Zambelli, T.; Zenobi, R.; Vorholt, J.A. Single-Cell Mass Spectrometry of Metabolites Extracted from Live Cells by Fluidic Force Microscopy. Anal. Chem. 2017, 89, 5017–5023. https://doi.org/10.1021/acs.analchem.7b00367.
- 30.
Saha, P.; Duanis-Assaf, T.; Reches, M. Fundamentals and Applications of FluidFM Technology in Single-Cell Studies. Adv. Mater. Interfaces 2020, 7, 2001115. https://doi.org/10.1002/admi.202001115.
- 31.
Sterling, H.J.; Batchelor, J.D.; Wemmer, D.E.; Williams, E.R. Effects of Buffer Loading for Electrospray Ionization Mass Spectrometry of a Noncovalent Protein Complex That Requires High Concentrations of Essential Salts. J. Am. Soc. Mass Spectrom. 2010, 21, 1045–1049. https://doi.org/10.1016/j.jasms.2010.02.003.
- 32.
Abouleila, Y.; Onidani, K.; Ali, A.; Shoji, H.; Kawai, T.; Lim, C.T.; Kumar, V.; Okaya, S.; Kato, K.; Hiyama, E.; et al. Live Single Cell Mass Spectrometry Reveals Cancer‐specific Metabolic Profiles of Circulating Tumor Cells. Cancer Sci. 2019, 110, 697–706. https://doi.org/10.1111/cas.13915.
- 33.
Sakata, J.; Furusho, A.; Sugiyama, E.; Sakane, I.; Todoroki, K.; Mizuno, H. Development of a Highly Efficient Solubilization Method for Mass Spectrometric Analysis of Phospholipids in Living Single Cells. Anal. Sci. 2024, 40, 917–924. https://doi.org/10.1007/s44211-024-00542-6.
- 34.
Yao, H.; Zhao, H.; Zhao, X.; Pan, X.; Feng, J.; Xu, F.; Zhang, S.; Zhang, X. Label-Free Mass Cytometry for Unveiling Cellular Metabolic Heterogeneity. Anal. Chem. 2019, 91, 9777–9783. https://doi.org/10.1021/acs.analchem.9b01419.
- 35.
Shao, Y.; Zhou, Y.; Liu, Y.; Zhang, W.; Zhu, G.; Zhao, Y.; Zhang, Q.; Yao, H.; Zhao, H.; Guo, G.; et al. Intact Living-Cell Electrolaunching Ionization Mass Spectrometry for Single-Cell Metabolomics. Chem. Sci. 2022, 13, 8065–8073. https://doi.org/10.1039/D2SC02569H.
- 36.
Qin, S.; Zhang, Y.; Shi, M.; Miao, D.; Lu, J.; Wen, L.; Bai, Y. In-Depth Organic Mass Cytometry Reveals Differential Contents of 3-Hydroxybutanoic Acid at the Single-Cell Level. Nat. Commun. 2024, 15, 4387. https://doi.org/10.1038/s41467-024-48865-2.
- 37.
Xu, S.; Xue, J.; Bai, Y.; Liu, H. High-Throughput Single-Cell Immunoassay in the Cellular Native Environment Using Online Desalting Dual-Spray Mass Spectrometry. Anal. Chem. 2020, 92, 15854–15861. https://doi.org/10.1021/acs.analchem.0c03167.
- 38.
Liu, H.; Wu, T.; He, H.; Zhou, R.; Zhao, J.; Zhan, L.; Hou, Z.; Huang, G. Dual Desalting Electrospray Strategy for In-Cell Mass Spectrometry to Reveal Novel Sphingolipid Metabolism in an Epithelial–Mesenchymal Transition. Anal. Chem. 2025, 97, 8337–8345. https://doi.org/10.1021/acs.analchem.4c06669.
- 39.
Zhang, W.; Li, N.; Lin, L.; Huang, Q.; Uchiyama, K.; Lin, J.-M. Concentrating Single Cells in Picoliter Droplets for Phospholipid Profiling on a Microfluidic System. Small 2020, 16, e1903402. https://doi.org/10.1002/smll.201903402.
- 40.
Li, Z.; Wang, Z.; Pan, J.; Ma, X.; Zhang, W.; Ouyang, Z. Single-Cell Mass Spectrometry Analysis of Metabolites Facilitated by Cell Electro-Migration and Electroporation. Anal. Chem. 2020, 92, 10138–10144. https://doi.org/10.1021/acs.analchem.0c02147.
- 41.
Ge, X.; Xing, X.; Wang, X.; Yin, Y.; Shen, X.; Ouyang, J.; Na, N. Glass Electrospray for Mass Spectrometry in Situ Detection of Living Cells. Chem. Commun. 2025, 61, 4164–4167. https://doi.org/10.1039/D4CC06657J.
- 42.
Zhuang, M.; Hou, Z.; Chen, P.; Liang, G.; Huang, G. Introducing Charge Tag via Click Reaction in Living Cells for Single Cell Mass Spectrometry. Chem. Sci. 2020, 11, 7308–7312. https://doi.org/10.1039/D0SC00259C.
- 43.
Fienberg, H.G.; Simonds, E.F.; Fantl, W.J.; Nolan, G.P.; Bodenmiller, B. A Platinum‐based Covalent Viability Reagent for Single‐cell Mass Cytometry. Cytom. Part A 2012, 81, 467–475. https://doi.org/10.1002/cyto.a.22067.
- 44.
Pandian, K.; Hetzel, L.A.; Zwier, R.; van Veldhuizen, P.; Schubert, C.; Karuppusamy, J.; Harms, A.C.; Ali, A.; Hankemeier, T. Enabling High-Sensitivity Live Single-Cell Mass Spectrometry Using an Integrated Electrical Lysis and Nano Electrospray Ionization Interface. Anal. Chim. Acta 2024, 1324, 343068. https://doi.org/10.1016/j.aca.2024.343068.
- 45.
Esaki, T.; Masujima, T. Fluorescence Probing Live Single-Cell Mass Spectrometry for Direct Analysis of Organelle Metabolism. Anal. Sci. 2015, 31, 1211–1213. https://doi.org/10.2116/analsci.31.1211.
- 46.
Xu, S.-T.; Yang, C.; Yan, X.-P. Organic Mass Cytometry Discriminating Cycle Stages of Single Cells with Small Molecular Indicators. Anal. Chem. 2023, 95, 2312–2320. https://doi.org/10.1021/acs.analchem.2c04165.
- 47.
Wang, Z.; Ge, S.; Liao, T.; Yuan, M.; Qian, W.; Chen, Q.; Liang, W.; Cheng, X.; Zhou, Q.; Ju, Z.; et al. Integrative Single-Cell Metabolomics and Phenotypic Profiling Reveals Metabolic Heterogeneity of Cellular Oxidation and Senescence. Nat. Commun. 2025, 16, 2740. https://doi.org/10.1038/s41467-025-57992-3.
- 48.
Zhang, Y.; Shi, M.; Li, M.; Qin, S.; Miao, D.; Bai, Y. Dynamic Single-Cell Metabolomics Reveals Cell-Cell Interaction between Tumor Cells and Macrophages. Nat. Commun. 2025, 16, 4582. https://doi.org/10.1038/s41467-025-59878-w.
- 49.
Shi, C.; Jia, H.; Chen, S.; Huang, J.; Peng, Y.; Guo, W. Hydrogen/Deuterium Exchange Aiding Metabolite Identification in Single-Cell Nanospray High-Resolution Mass Spectrometry Analysis. Anal. Chem. 2022, 94, 650–657. https://doi.org/10.1021/acs.analchem.1c02057.
- 50.
Sun, M.; Yang, Z. Metabolomic Studies of Live Single Cancer Stem Cells Using Mass Spectrometry. Anal. Chem. 2019, 91, 2384–2391. https://doi.org/10.1021/acs.analchem.8b05166.
- 51.
Xu, Y.; Hu, X.; Yuan, Y.; Liu, W.; Wang, J.; Yang, C.; Shi, X.; Qin, W.; Wen, L.; Lin, M.; et al. Prediction of Lung Cancer Metastasis Risk Based on Single-Cell Metabolic Profiling of Circulating Tumor Cells. Adv. Sci. 2025, 12, e08878. https://doi.org/10.1002/advs.202508878.
- 52.
Pan, N.; Standke, S.J.; Kothapalli, N.R.; Sun, M.; Bensen, R.C.; Burgett, A.W.G.; Yang, Z. Quantification of Drug Molecules in Live Single Cells Using the Single-Probe Mass Spectrometry Technique. Anal. Chem. 2019, 91, 9018–9024. https://doi.org/10.1021/acs.analchem.9b01311.
- 53.
Zhu, Y.; Liu, R.; Yang, Z. Redesigning the T-Probe for Mass Spectrometry Analysis of Online Lysis of Non-Adherent Single Cells. Anal. Chim. Acta 2019, 1084, 53–59. https://doi.org/10.1016/j.aca.2019.07.059.
- 54.
Chen, X.; Peng, Z.; Yang, Z. Metabolomics Studies of Cell-Cell Interactions Using Single Cell Mass Spectrometry Combined with Fluorescence Microscopy. Chem. Sci. 2022, 13, 6687–6695. https://doi.org/10.1039/d2sc02298b.
- 55.
Liu, R.; Pan, N.; Zhu, Y.; Yang, Z. T-Probe: An Integrated Microscale Device for Online in situ Single Cell Analysis and Metabolic Profiling Using Mass Spectrometry. Anal. Chem. 2018, 90, 11078–11085. https://doi.org/10.1021/acs.analchem.8b02927.
- 56.
Shen, Z.; Zhao, H.; Yao, H.; Pan, X.; Yang, J.; Zhang, S.; Han, G.; Zhang, X. Dynamic Metabolic Change of Cancer Cells Induced by Natural Killer Cells at the Single-Cell Level Studied by Label-Free Mass Cytometry. Chem. Sci. 2022, 13, 1641–1647. https://doi.org/10.1039/D1SC06366A.
- 57.
Pan, S.; Liu, C.; Yao, H.; Pan, X.; Li, J.; Yang, J.; Du, M.; Liu, P.; Zhang, S.; Zhang, X. Single-Cell Metabolite Profiling Enables Information-Rich Classification of Lymphocyte Types and Subtypes. Chem. Commun. 2024, 60, 392–395. https://doi.org/10.1039/D3CC05011D.
- 58.
Hu, X.; Liu, X.; Feng, D.; Xu, T.; Li, H.; Hu, C.; Wang, Z.; Liu, X.; Yin, P.; Shi, X.; et al. Polarization of Macrophages in Tumor Microenvironment Using High-Throughput Single-Cell Metabolomics. Anal. Chem. 2024, 96, 14935–14943. https://doi.org/10.1021/acs.analchem.4c02989.
- 59.
Shimizu, T.; Miyakawa, S.; Esaki, T.; Mizuno, H.; Masujima, T.; Koshiba, T.; Seo, M. Live Single-Cell Plant Hormone Analysis by Video-Mass Spectrometry. Plant Cell Physiol. 2015, 56, 1287–1296. https://doi.org/10.1093/pcp/pcv042.
- 60.
Lorenzo Tejedor, M.; Mizuno, H.; Tsuyama, N.; Harada, T.; Masujima, T. In Situ Molecular Analysis of Plant Tissues by Live Single-Cell Mass Spectrometry. Anal Chem 2012, 84, 5221–5228. https://doi.org/10.1021/ac202447t.
- 61.
Han, J.; Fan, D.-J.; Jiang, L.-C.; Lu, Z.; Ye, T.; Wei, Z.; Feng, Y.-Q. In Situ Dynamic Molecular Monitoring of Single Seed Exudates by Induced Electrospray Ionization Mass Spectrometry. Anal. Chem. 2025, 97, 13912–13921. https://doi.org/10.1021/acs.analchem.5c01437.
- 62.
Onjiko, R.M.; Morris, S.E.; Moody, S.A.; Nemes, P. Single-Cell Mass Spectrometry with Multi-Solvent Extraction Identifies Metabolic Differences between Left and Right Blastomeres in the 8-Cell Frog (Xenopus) Embryo. Analyst 2016, 141, 3648–3656. https://doi.org/10.1039/C6AN00200E.
- 63.
Onjiko, R.M.; Plotnick, D.O.; Moody, S.A.; Nemes, P. Metabolic Comparison of Dorsal versus Ventral Cells Directly in the Live 8-Cell Frog Embryo by Microprobe Single-Cell CE-ESI-MS. Anal. Methods 2017, 9, 4964–4970. https://doi.org/10.1039/C7AY00834A.
- 64.
Li, G.; Yuan, S.; Zheng, S.; Liu, Y.; Huang, G. In Situ Living Cell Protein Analysis by Single-Step Mass Spectrometry. Anal. Chem. 2018, 90, 3409–3415. https://doi.org/10.1021/acs.analchem.7b05055.
- 65.
Li, G.; Yuan, S.; Pan, Y.; Liu, Y.; Huang, G. Binding States of Protein–Metal Complexes in Cells. Anal. Chem. 2016, 88, 10860–10866. https://doi.org/10.1021/acs.analchem.6b00032.
- 66.
Zhan, L.; Hou, Z.; Wang, Y.; Liu, H.; Liu, Y.; Huang, G. Rapid Profiling of Metabolic Perturbations to Antibiotics in Living Bacteria by Induced Electrospray Ionization Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2022, 33, 1960–1966. https://doi.org/10.1021/jasms.2c00199.
- 67.
Amantonico, A.; Urban, P.L.; Fagerer, S.R.; Balabin, R.M.; Zenobi, R. Single-Cell MALDI-MS as an Analytical Tool for Studying Intrapopulation Metabolic Heterogeneity of Unicellular Organisms. Anal. Chem. 2010, 82, 7394–7400. https://doi.org/10.1021/ac1015326.
- 68.
Baumeister, T.U.H.; Vallet, M.; Kaftan, F.; Svatoš, A.; Pohnert, G. Live Single-Cell Metabolomics with Matrix-Free Laser/Desorption Ionization Mass Spectrometry to Address Microalgal Physiology. Front. Plant Sci. 2019, 10, 172. https://doi.org/10.3389/fpls.2019.00172.
- 69.
Yao, H.; Yang, J.; Wang, Z.; Pan, X.; Pan, J.; Li, H.; Zhang, S. High-Throughput Metabolite Analysis of Unicellular Microalgae by Orthogonal Hybrid Ionization Label-Free Mass Cytometry. Anal. Chem. 2024, 96, 11404–11411. https://doi.org/10.1021/acs.analchem.4c01541.
- 70.
Xue, S.; Qiao, Y.; Jia, H.; Yang, Y.; Shi, L.; Zhang, Z.; Peng, Y.; Guo, W. Discrimination of Microalgal Cellular Metabolic Heterogeneity and 13C Metabolic Flux Analysis via Micro Label-Free Mass Cytometry. Anal. Chem. 2025, 97, 18382–18391. https://doi.org/10.1021/acs.analchem.5c03905.

This work is licensed under a Creative Commons Attribution 4.0 International License.


