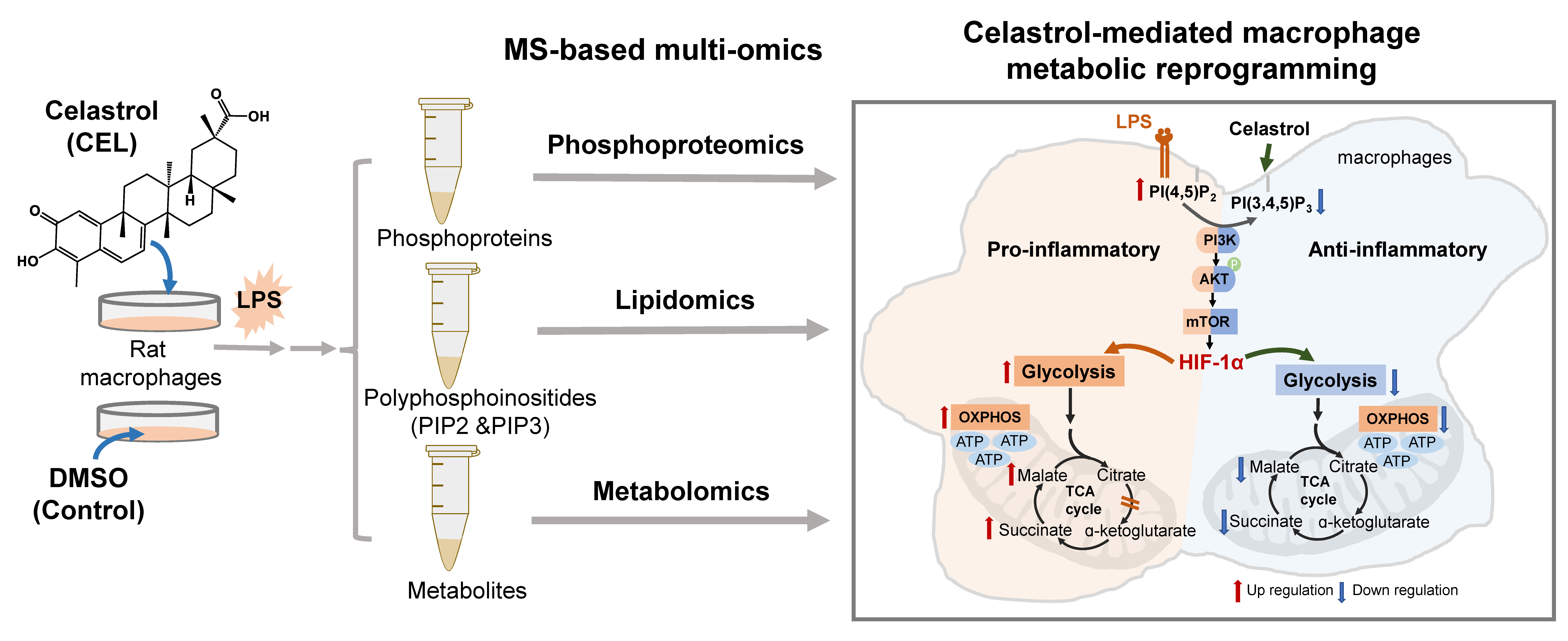
Celastrol (CEL), a bioactive compound derived from Tripterygium wilfordii, exerts potent anti-inflammatory and metabolic-regulating properties, though its underlying mechanisms remain incompletely elucidated. Herein, we combined phosphoproteomics, targeted lipidomics, and metabolomics to delineate how CEL reprograms macrophage metabolism to resolve inflammation. Temporal phosphoproteomic analysis in LPS-stimulated macrophages revealed that CEL dynamically suppressed phosphorylation events across inflammatory pathways, with the PI3K/AKT/mTOR axis emerging as a central regulated pathway. We successfully quantified 18 PIP2 and 14 PIP3 species by shotgun lipidomics and found that CEL significantly reduced PIP3/PIP2 ratios, inhibiting PI3K activity and attenuating mTOR-dependent phosphorylation of downstream effectors. We further found that the suppression of PI3K disrupted HIF-1α stabilization and nuclear translocation, reversing LPS-driven metabolic shifts: CEL dampened glycolysis while restoring oxidative phosphorylation (OXPHOS) and rectifying TCA cycle fragmentation, as evidenced by reduced succinate/α-ketoglutarate ratios. CEL skewed macrophages toward an anti-inflammatory M2 phenotype, marked by downregulated CD86 and upregulated CD163. Overall, we unveil HIF-1α as a critical mediator of CEL’s anti-inflammatory effect, bridging phosphoprotein signaling rewiring to metabolic reprogramming. This mechanistic insight positions CEL as a therapeutic candidate for inflammatory diseases via metabolic modulation.
- Open Access
- Article
Multi-Omics Analysis Reveals Signaling-Metabolic Reprogramming Crosstalk during Celastrol-Mediated Inflammatory Response
- Guangshan Xie 1,2,
- Lin Zhu 2,*,
- Xin Diao 2,
- Xiuli Su 3,
- Li Zhong 2,
- Zongwei Cai 2,3,*
Author Information
Received: 24 Jul 2025 | Revised: 11 Sep 2025 | Accepted: 11 Oct 2025 | Published: 12 Jan 2026
Abstract
Graphical Abstract
Keywords
multi-omics analysis | celastrol | signaling pathway | metabolic reprogramming | inflammatory response
References
- 1.
Liu, J.; Lee, J.; Hernandez, M.A.S.; Mazitschek, R.; Ozcan, U. Treatment of Obesity with Celastrol. Cell 2015, 161, 999–1011.
- 2.
An, L.; Li, Z.; Shi, L.; Wang, L.; Wang, Y.; Jin, L.; Shuai, X.; Li, J. Inflammation-Targeted Celastrol Nanodrug Attenuates Collagen-Induced Arthritis through NF-ΚB and Notch1 Pathways. Nano Lett. 2020, 20, 7728–7736.
- 3.
Ma, X.; Xu, L.; Alberobello, A.T.; Gavrilova, O.; Bagattin, A.; Skarulis, M.; Liu, J.; Finkel, T.; Mueller, E. Celastrol Protects against Obesity and Metabolic Dysfunction through Activation of a HSF1-PGC1α Transcriptional Axis. Cell Metab. 2015, 22, 695–708.
- 4.
Cascão, R.; Vidal, B.; Raquel, H.; Neves-Costa, A.; Figueiredo, N.; Gupta, V.; Fonseca, J.E.; Moita, L.F. Effective Treatment of Rat Adjuvant-Induced Arthritis by Celastrol. Autoimmun. Rev. 2012, 11, 856–862.
- 5.
Luo, D.; Guo, Y.; Cheng, Y.; Zhao, J.; Wang, Y.; Rong, J. Natural Product Celastrol Suppressed Macrophage M1 Polarization against Inflammation in Diet-Induced Obese Mice via Regulating Nrf2/HO−1, MAP Kinase and NF-ΚB Pathways. Aging 2017, 9, 2068–2081.
- 6.
Park, M.D.; Silvin, A.; Ginhoux, F.; Merad, M. Macrophages in Health and Disease. Cell 2022, 185, 4259.
- 7.
Russell, D.G.; Huang, L.; VanderVen, B.C. Immunometabolism at the Interface between Macrophages and Pathogens. Nat. Rev. Immunol. 2019, 19, 291–304.
- 8.
Navegantes, K.C.; Souza Gomes, R.; Pereira, P.A.T.; Czaikoski, P.G.; Azevedo, C.H.M.; Monteiro, M.C. Immune Modulation of Some Autoimmune Diseases: The Critical Role of Macrophages and Neutrophils in the Innate and Adaptive Immunity. J. Transl. Med. 2017, 15, 36.
- 9.
Liu, P.S.; Wang, H.; Li, X.; Chao, T.; Teav, T.; Christen, S.; DI Conza, G.; Cheng, W.C.; Chou, C.H.; Vavakova, M.; et al. α-Ketoglutarate Orchestrates Macrophage Activation through Metabolic and Epigenetic Reprogramming. Nat. Immunol. 2017, 18, 985–994.
- 10.
Putnam, K.; Shoemaker, R.; Yiannikouris, F.; Cassis, L.A. The Renin-Angiotensin System: A Target of and Contributor to Dyslipidemias, Altered Glucose Homeostasis, and Hypertension of the Metabolic Syndrome. Am. J. Physiol. Heart Circ. Physiol. 2012, 302, H1219–H1230.
- 11.
Pearce, E.L.; Pearce, E.J. Metabolic Pathways in Immune Cell Activation and Quiescence. Immunity 2013, 38, 633–643.
- 12.
Cheng, S.C.; Quintin, J.; Cramer, R.A.; Shepardson, K.M.; Saeed, S.; Kumar, V.; Giamarellos-Bourboulis, E.J.; Martens, J.H.A.; Rao, N.A.; Aghajanirefah, A.; et al. MTOR- and HIF-1α-Mediated Aerobic Glycolysis as Metabolic Basis for Trained Immunity. Science 2014, 345, 1250684.
- 13.
Needham, E.J.; Parker, B.L.; Burykin, T.; James, D.E.; Humphrey, S.J. Illuminating the Dark Phosphoproteome. Sci. Signal. 2019, 12, eaau8645.
- 14.
Liu, S.; Zhu, L.; Xie, G.; Mok, B.W.Y.; Yang, Z.; Deng, S.; Lau, S.Y.; Chen, P.; Wang, P.; Chen, H.; et al. Potential Antiviral Target for SARS-CoV-2: A Key Early Responsive Kinase during Viral Entry. CCS Chem. 2022, 4, 112–121.
- 15.
Xie, G.; Zhu, L.; Song, Y.; Huang, W.; Hu, D.; Cai, Z. An Integrated Quantitative Proteomics Strategy Reveals the Dual Mechanisms of Celastrol against Acute Inflammation. Chin. Chem. Lett. 2021, 32, 2164–2168.
- 16.
Xie, G.; Zhu, L.; Liu, S.; Li, C.; Diao, X.; Zhang, Y.; Su, X.; Song, Y.; Cao, G.; Zhong, L.; et al. Multi-Omics Analysis of Attenuated Variant Reveals Potential Evaluation Marker of Host Damaging for SARS-CoV-2 Variants. Sci. China Life Sci. 2024, 67, 83–95.
- 17.
Zhao, J.; Liu, H.; Chen, Q.; Xia, M.; Wan, L.; Yu, W.; Liu, C.; Hao, X.; Tang, C.; Chen, G.; et al. Mechanistic Study of Celastrol-Mediated Inhibition of Proinflammatory Activation of Macrophages in IgA Nephropathy via down-Regulating ECM1. Int. J. Biol. Sci. 2024, 20, 5731–5746.
- 18.
Shirai, T.; Nakai, A.; Ando, E.; Fujimoto, J.; Leach, S.; Arimori, T.; Higo, D.; van Eerden, F.J.; Tulyeu, J.; Liu, Y.C.; et al. Celastrol Suppresses Humoral Immune Responses and Autoimmunity by Targeting the COMMD3/8 Complex. Sci. Immunol. 2023, 8, eadc9324.
- 19.
Chang-Lun, H.; Der-Yuan, C.; Chih-Chen, T.; Jhen-Wei, L.; Bor-Show, T.; Tsai-Ching, H. Celastrol Attenuates Human Parvovirus B19 NS1-induced NLRP3 Inflammasome Activation in Macrophages. Mol. Med. Rep. 2023, 28, 193.
- 20.
Venkatesha, S.H.; Dudics, S.; Astry, B.; Moudgil, K.D. Control of Autoimmune Inflammation by Celastrol, a Natural Triterpenoid. Pathog. Dis. 2016, 74, ftw059.
- 21.
Wang, S.; Huang, Z.; Lei, Y.; Han, X.; Tian, D.; Gong, J.; Liu, M. Celastrol Alleviates Autoimmune Hepatitis Through the PI3K/AKT Signaling Pathway Based on Network Pharmacology and Experiments. Front. Pharmacol. 2022, 13, 816350.
- 22.
Nakedi, K.C.; Calder, B.; Banerjee, M.; Giddey, A.; Nel, A.J.M.; Garnett, S.; Blackburn, J.M.; Soares, N.C. Identification of Novel Physiological Substrates of Mycobacterium Bovis BCG Protein Kinase G (PknG) by Label-Free Quantitative Phosphoproteomics. Mol. Cell. Proteom. 2018, 17, 1365.
- 23.
Lien, E.C.; Lyssiotis, C.A.; Cantley, L.C. Metabolic Reprogramming by the PI3K-Akt-MTOR Pathway in Cancer. Recent Results Cancer Res. 2016, 207, 39–72.
- 24.
Steelman, L.S.; Chappell, W.H.; Abrams, S.L.; Kempf, C.R.; Long, J.; Laidler, P.; Mijatovic, S.; Maksimovic-Ivanic, D.; Stivala, F.; Mazzarino, M.C.; et al. Roles of the Raf/MEK/ERK and PI3K/PTEN/Akt/MTOR Pathways in Controlling Growth and Sensitivity to Therapy-Implications for Cancer and Aging. Aging 2011, 3, 192.
- 25.
Kuo, W.L.; Sharifi, M.N.; Lingen, M.W.; Ahmed, O.; Liu, J.; Nagilla, M.; Macleod, K.F.; Cohen, E.E.W. P62/SQSTM1 Accumulation in Squamous Cell Carcinoma of Head and Neck Predicts Sensitivity to Phosphatidylinositol 3-Kinase Pathway Inhibitors. PLoS ONE 2014, 9, e90171.
- 26.
Wang, C.; Palavicini, J.P.; Wang, M.; Chen, L.; Yang, K.; Crawford, P.A.; Han, X. Comprehensive and Quantitative Analysis of Polyphosphoinositide Species by Shotgun Lipidomics Revealed Their Alterations in Db/Db Mouse Brain. Anal. Chem. 2016, 88, 12137–12144.
- 27.
Arsham, A.M.; Plas, D.R.; Thompson, C.B.; Celeste Simon, M. Phosphatidylinositol 3-Kinase/Akt Signaling Is Neither Required for Hypoxic Stabilization of HIF-1 Alpha nor Sufficient for HIF-1-Dependent Target Gene Transcription. J. Biol. Chem. 2002, 277, 15162–15170.
- 28.
Taylor, C.T.; Scholz, C.C. The Effect of HIF on Metabolism and Immunity. Nat. Rev. Nephrol. 2022, 18, 573–587.
- 29.
Mylonis, I.; Chachami, G.; Samiotaki, M.; Panayotou, G.; Paraskeva, E.; Kalousi, A.; Georgatsou, E.; Bonanou, S.; Simos, G. Identification of MAPK Phosphorylation Sites and Their Role in the Localization and Activity of Hypoxia-Inducible Factor-1alpha. J. Biol. Chem. 2006, 281, 33095–33106.
- 30.
Tannahill, G.M.; Curtis, A.M.; Adamik, J.; Palsson-Mcdermott, E.M.; McGettrick, A.F.; Goel, G.; Frezza, C.; Bernard, N.J.; Kelly, B.; Foley, N.H.; et al. Succinate Is a Danger Signal That Induces IL-1β via HIF-1α. Nature 2013, 496, 238.
- 31.
Lian, G.; Li, X.; Zhang, L.; Zhang, Y.; Sun, L.; Zhang, X.; Liu, H.; Pang, Y.; Kong, W.; Zhang, T.; et al. Macrophage Metabolic Reprogramming Aggravates Aortic Dissection through the HIF1α-ADAM17 Pathway✰. EBioMedicine 2019, 49, 291–304.
- 32.
Dang, B.; Gao, Q.; Zhang, L.; Zhang, J.; Cai, H.; Zhu, Y.; Zhong, Q.; Liu, J.; Niu, Y.; Mao, K.; et al. The Glycolysis/HIF-1α Axis Defines the Inflammatory Role of IL-4-Primed Macrophages. Cell Rep. 2023, 42, 112471.
- 33.
Jha, A.K.; Huang, S.C.C.; Sergushichev, A.; Lampropoulou, V.; Ivanova, Y.; Loginicheva, E.; Chmielewski, K.; Stewart, K.M.; Ashall, J.; Everts, B.; et al. Network Integration of Parallel Metabolic and Transcriptional Data Reveals Metabolic Modules That Regulate Macrophage Polarization. Immunity 2015, 42, 419–430.
- 34.
Metallo, C.M.; Gameiro, P.A.; Bell, E.L.; Mattaini, K.R.; Yang, J.; Hiller, K.; Jewell, C.M.; Johnson, Z.R.; Irvine, D.J.; Guarente, L.; et al. Reductive Glutamine Metabolism by IDH1 Mediates Lipogenesis under Hypoxia. Nature 2011, 481, 380–384.
- 35.
Lee, J.H.; Koo, T.H.; Yoon, H.; Jung, H.S.; Jin, H.Z.; Lee, K.; Hong, Y.S.; Lee, J.J. Inhibition of NF-ΚB Activation through Targeting IκB Kinase by Celastrol, a Quinone Methide Triterpenoid. Biochem. Pharmacol. 2006, 72, 1311–1321.
- 36.
Zhao, J.; Sun, Y.; Shi, P.; Dong, J.N.; Zuo, L.G.; Wang, H.G.; Gong, J.F.; Li, Y.; Gu, L.L.; Li, N.; et al. Celastrol Ameliorates Experimental Colitis in IL-10 Deficient Mice via the up-Regulation of Autophagy. Int. Immunopharmacol. 2015, 26, 221–228.
- 37.
Ersahin, T.; Tuncbag, N.; Cetin-Atalay, R. The PI3K/AKT/MTOR Interactive Pathway. Mol. Biosyst. 2015, 11, 1946–1954.
- 38.
Li, W.; Tang, X.; Zheng, Y.; Xu, X.; Zhao, N.; Tsao, B.P.; Feng, X.; Sun, L. Phosphatidic Acid Promoting the Generation of Interleukin-17A Producing Double-Negative T Cells by Enhancing MTORC1 Signaling in Lupus. Arthritis Rheumatol. 2024, 76, 1096–1108.
- 39.
Jiang, S.; Yang, H.; Li, M. Emerging Roles of Lysophosphatidic Acid in Macrophages and Inflammatory Diseases. Int. J. Mol. Sci. 2023, 24, 12524.
- 40.
Choy, C.H.; Han, B.K.; Botelho, R.J. Phosphoinositide Diversity, Distribution, and Effector Function: Stepping Out of the Box. Bioessays 2017, 39, 12524.
- 41.
Shulga, Y.V.; Anderson, R.A.; Topham, M.K.; Epand, R.M. Phosphatidylinositol-4-Phosphate 5-Kinase Isoforms Exhibit Acyl Chain Selectivity for Both Substrate and Lipid Activator. J. Biol. Chem. 2012, 287, 35953–35963.
- 42.
Schmid, A.C.; Wise, H.M.; Mitchell, C.A.; Nussbaum, R.; Woscholski, R. Type II Phosphoinositide 5-Phosphatases Have Unique Sensitivities towards Fatty Acid Composition and Head Group Phosphorylation. FEBS Lett. 2004, 576, 9–13.
- 43.
Barneda, D.; Janardan, V.; Niewczas, I.; Collins, D.M.; Cosulich, S.; Clark, J.; Stephens, L.R.; Hawkins, P.T. Acyl Chain Selection Couples the Consumption and Synthesis of Phosphoinositides. EMBO J. 2022, 41, e110038.
- 44.
Kim, Y.J.; Sengupta, N.; Sohn, M.; Mandal, A.; Pemberton, J.G.; Choi, U.; Balla, T. Metabolic Routing Maintains the Unique Fatty Acid Composition of Phosphoinositides. EMBO Rep. 2022, 23, e54532.
- 45.
Michell, R.H. Inositol Phospholipids and Cell Surface Receptor Function. BBA—Rev. Biomembr. 1975, 415, 81–147.
- 46.
Manni, M.M.; Tiberti, M.L.; Pagnotta, S.; Barelli, H.; Gautier, R.; Antonny, B. Acyl Chain Asymmetry and Polyunsaturation of Brain Phospholipids Facilitate Membrane Vesiculation without Leakage. Elife 2018, 7, e34394.
- 47.
Albanese, A.; Daly, L.A.; Mennerich, D.; Kietzmann, T.; Sée, V. The Role of Hypoxia-Inducible Factor Post-Translational Modifications in Regulating Its Localisation, Stability, and Activity. Int. J. Mol. Sci. 2020, 22, 268.
- 48.
Zhou, J.; Fandrey, J.; Schümann, J.; Tiegs, G.; Brüne, B. NO and TNF-Alpha Released from Activated Macrophages Stabilize HIF-1alpha in Resting Tubular LLC-PK1 Cells. Am. J. Physiol. Cell Physiol. 2003, 284, C439–C46
- 49.
Corcoran, S.E.; O’Neill, L.A.J. HIF1α and Metabolic Reprogramming in Inflammation. J. Clin. Investig. 2016, 126, 3699–3707.
- 50.
Huang, L.; Zhang, Z.; Zhang, S.; Ren, J.; Zhang, R.; Zeng, H.; Li, Q.; Wu, G. Inhibitory Action of Celastrol on Hypoxia-Mediated Angiogenesis and Metastasis via the HIF-1α Pathway. Int. J. Mol. Med. 2011, 27, 407–415.
- 51.
Palmieri, E.M.; Gonzalez-Cotto, M.; Baseler, W.A.; Davies, L.C.; Ghesquière, B.; Maio, N.; Rice, C.M.; Rouault, T.A.; Cassel, T.; Higashi, R.M.; et al. Nitric Oxide Orchestrates Metabolic Rewiring in M1 Macrophages by Targeting Aconitase 2 and Pyruvate Dehydrogenase. Nat. Commun. 2020, 11, 698
- 52.
Li, Y.; Li, Y.C.; Liu, X.T.; Zhang, L.; Chen, Y.H.; Zhao, Q.; Gao, W.; Liu, B.; Yang, H.; Li, P. Blockage of Citrate Export Prevents TCA Cycle Fragmentation via Irg1 Inactivation. Cell Rep. 2022, 38, 110391.
- 53.
Joo, H.Y.; Jung, J.K.; Kim, M.Y.; Woo, S.R.; Jeong, J.M.; Park, E.R.; Kim, Y.M.; Park, J.J.; Kim, J.; Yun, M.; et al. NADH Elevation during Chronic Hypoxia Leads to VHL-Mediated HIF-1α Degradation via SIRT1 Inhibition. Cell Biosci. 2023, 13, 182.
- 54.
Cramer, T.; Yamanishi, Y.; Clausen, B.E.; Förster, I.; Pawlinski, R.; Mackman, N.; Haase, V.H.; Jaenisch, R.; Corr, M.; Nizet, V.; et al. HIF-1α Is Essential for Myeloid Cell-Mediated Inflammation. Cell 2003, 112, 645–657.
- 55.
Kelly, B.; O’Neill, L.A.J. Metabolic Reprogramming in Macrophages and Dendritic Cells in Innate Immunity. Cell Res. 2015, 25, 771–784.
- 56.
Li, W.; Cai, Z.N.; Mehmood, S.; Liang, L.L.; Liu, Y.; Zhang, H.Y.; Chen, Y.; Lu, Y.M. Anti-Inflammatory Effects of Morchella Esculenta Polysaccharide and Its Derivatives in Fine Particulate Matter-Treated NR8383 Cells. Int. J. Biol. Macromol. 2019, 129, 904–915.
- 57.
Wong, L.Y.F.; Cheung, B.M.Y.; Li, Y.Y.; Tang, F. Adrenomedullin Is Both Proinflammatory and Antiinflammatory: Its Effects on Gene Expression and Secretion of Cytokines and Macrophage Migration Inhibitory Factor in NR8383 Macrophage Cell Line. Endocrinology 2005, 146, 1321–1327.
- 58.
Chen, S.; Hu, Y.; Zhang, J.; Zhang, P. Anti-inflammatory Effect of Salusin-β Knockdown on LPS-activated Alveolar Macrophages via NF-κB Inhibition and HO-1 Activation. Mol. Med. Rep. 2021, 23, 127.

This work is licensed under a Creative Commons Attribution 4.0 International License.


