Downloads
Download
Additional Files
Download - Supplementary file


This work is licensed under a Creative Commons Attribution 4.0 International License.
Review
Progress of 3D Organoid Technology for Preclinical Investigations: Towards Human In Vitro Models
Yingjuan Liu *, Honglin Xu, Sabu Abraham, Xin Wang, and Bernard D. Keavney*
Division of Cardiovascular Sciences, Faculty of Biology, Medicine and Health, University of Manchester, M13 9PT, UK.
* Correspondence: yingjuan.liu@manchester.ac.uk (Yingjuan Liu);
bernard.keavney@manchester.ac.uk (Bernard D. Keavney)
Received: 1 November 2022
Accepted: 30 November 2022
Published: 21 December 2022
Abstract: Currently, with an increased requirement for new therapeutic strategies, preclinical drug testing or screening platforms have rapidly evolved in recent years. In comparison to traditional 2D cell cultures, 3D organoids or spheroids with or without scaffolds improve the microenvironment of in vitro cultures, advancing the in vitro biological observation and enabling mechanistic studies of drug reactions in the human tissue-like environment. 3D organoids and spheroids are straightforward to produce, and relatively uniform in size and shape. This helps to facilitate high throughput screening requirements. Spheroids and organoids have been applied in anti-cancer drug testing, toxicity evaluations, as well as mechanism studies for variable organ systems, including the intestine, liver, pancreas, brain, and heart. Among 3D cultures of spheroids and organoids, ‘tumour spheroids’ formed by dissociated tumour tissues or cancer cell lines are relatively simple in composition and commonly applied to anticancer drug screening. The ‘healthy organoids’ differentiated from hiPSCs/hESCs are more complex in cell composition, distribution, structure and function with higher similarity to in vivo organs, and have found applications in toxicity tests, personalised medicine, and therapeutic and mechanistic studies. In most cases, the multicellular 3D organoids are more resistant and stable in reaction to stimulations or chemicals in vitro , suggesting more accurate modelling of in vivo responses. Here, we review recent progress in human-origin organoid/spheroid systems and their applications in preclinical studies.
Keywords:
drug screening disease modelling preclinical application self-assembly self-organising 3D culture1.Introduction
In developing novel drugs, preclinical drug screening and toxicity tests are indispensable. While animal testing is the gold standard, efforts by regulatory bodies to reduce the use of animals, and lack of perfect human correspondence of animal testing platforms, mandate the development and use of more sophisticated cellular models and strategies [1]. Among cellular models, 2D monolayer cell cultures have been applied for drug tests, screening and mechanism investigations for a long time. However, in general, cell biological reactions at 2D levels do not recapitulate in vivo tissue properties, absorption kinetics, and functional responses [2]. For example, recent studies comparing the pharmacological effects of anticancer drugs in 2D and 3D models show disagreements in reactive cell sensitivities, viabilities, and apoptosis rates [3–5]. In most cases, the 3D models, which exhibit higher similarity at the transcriptional level with in vivo situations [6], are relatively more resistant to the cytotoxicity of drugs than monolayer cell cultures [7]. Cell matrix, hypoxia, structures and pharmacological dynamics are all possible factors that influence the differences [7]. Unlike 2D cultures, 3D models provide more cell-cell contacts, cell-microenvironment interactions, higher cell density and chemical exposure gradients within the ‘3D tissues’ [8], which are closer to the characteristics of in vivo tissues.
Among various 3D cultures, ‘organoids’ are ideal test platforms for pharmacological and toxicological evaluations due to their scaling-up potential. However, there is not yet a clear and standardised definition of ‘organoid’. The notion of ‘organoid’ is commonly mixed with ‘spheroid’. To provide a clear definition of both items and compare their application efficacy in drug screening and toxicity tests, we reviewed studies using 3D models of claimed ‘organoid’ or ‘spheroid’ with human cell origins in their investigations of preclinical drug tests or screening.
2.The ‘Organoid’ and ‘Spheroid’: Terminology
The first attempts to generate organoids were dissociation-reaggregation experiments in suspension culture from the 1900s [9]. Enlightening by matrix application in cell culture and stem cell differentiation [10–12], Sato et al. [13] in 2009 generated a self-organised intestinal organoid with functional structures. Following the success in generating intestinal organoids, gastric organoids, liver and pancreatic organoids, kidney organoids, brain and other organoids were gradually created and improved over the recent years [14].
Although there is not yet a standard definition of an organoid, it is generally conceptualised as a 3D multiple cellular cluster resembling the in vivo tissues and organs in structures, functionalities and mechanisms, with the capacity of self-renewing and self-organization [15,16]. ‘Organoids’ can be generated via self-assembly or scaffolding from multiple cell origins, including patient-derived cells, established cell lines, hiPSCs/hESCs, and hiPSC/hESC-derived cells with or without matrix. The organoids can display well-constructed spheres, compacted spheres, elongated spheres, loose spheres, branched spheres, and grape-like or stellar-like tissues [17]. Due to the shared sphere shape, it is necessary to apply criteria distinguishing between organoids and spheroids to avoid confusion. Organoids and spheroids overlap in specific cellular sources and culture conditions [18]. Spheroids are simpler cell aggregates of single or multiple cell types with relatively consistent cell distributions across the whole structure, retaining cell properties and reactivity to stimulations. The application of spheroids started even earlier than organoids, around the 1970s, when they were reaggregated from disassociated tumour cells in recapturing the functionality of tumour tissues [19,20]. Unlike spheroids, organoids are more complex in cell composition, functions, and structures, with either structuralised compartments, separated cell layers, or polarised cell distributions, like a miniature organ [21].
Since there is an increasing demand for 3D models in developing or screening new drugs or therapy methods, investigating pharmacological mechanisms and promoting personalised medicine, many different approaches of generating or crafting these 3D organoids or spheroids have been developed to address this problem.
3.Methods of Generating ‘Organoid’ and ‘Spheroid’
3.1.Cell Sources
‘Organoid’ and ‘Spheroid’ are different in functions, structures and compositions; They overlap in the cell sources used for construction. The general cell sources are derived from traditional 2D cultured cell lines, primary cell cultures or fresh dissociated tissues from patients (including adult-derived stem cells), hiPSCs and hiPSC-differentiated cells. Based on the cell source properties, the ‘organoid’ and ‘spheroid’ can be classified as ‘healthy cell derived’ and ‘tumour cell derived’ organoids or spheroids.
To understand the most recent distributions of cell sources applied in the production of spheroid and organoids, we did a sampling literature search in PubMed with the keywords of “organoid” or “spheroid” with “drug screening” or “drug test” in human studies during 2021–2022 (dataset in ‘Supplementary file’). Among the 49 studies identified, 41 use tumour cell-derived cultures in their investigations. Only eight studies prepare healthy cell-derived organoids or spheroids for screening or testing; six develop their screening models from hiPSCs, while the other two use patient-derived cells or immortalised cell lines.
In the majority of studies, tumour cell-derived cultures are named spheroids. Only six of the 42 tumour studies claimed the models as ‘tumour organoids’, ‘patient-derived organoids’ or ‘spheroidal 3D organoids’. Despite the asserted names of the cultures, all tumour cell-derived cultures are relatively simple in composition, originating from a single source of immortalised tumour cell lines or dissociated patient tumour tissues with or without supportive cell populations like fibroblast cells. Typically, these models keep the cell proliferation properties and can be passaged for expansion.
Healthy cell-derived cultures are mainly named ‘organoids’ and differentiated from hiPSCs. After the differentiation and induction of organoids, the cells are typically unable to be further passaged for expansion. In function, the healthy cell-derived cultures are more similar to healthy tissues in textures, structures, and functionalities. There are also studies including immortalised healthy cell lines, iPSC-differentiated cells, and primary tissue-derived cells in 3D cultures. In these circumstances, the constructions of these models are not as complex as the stem cell-derived organoids. These simpler 3D cultures are typically named ‘spheroid’.
3.2.‘Organoid’ and ‘Spheroid’ Culture
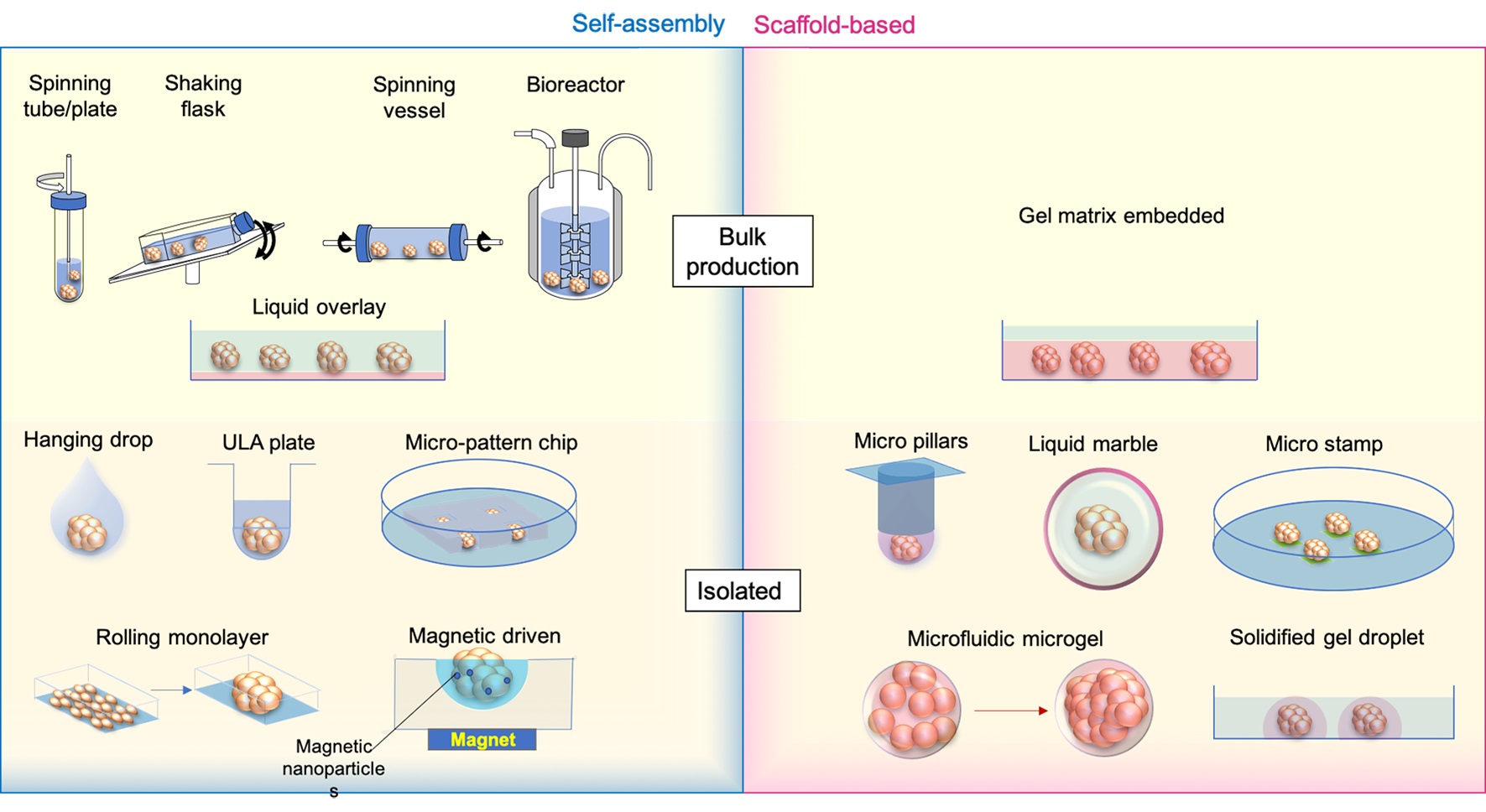
There are different ways of generating an ‘organoid’ or ‘spheroid’. Based on the methods of cell aggregation, the ‘organoid’ or ‘spheroid’ cultures are grouped as self-assembly cultures without scaffolds and scaffold-based cultures (Figure 1).
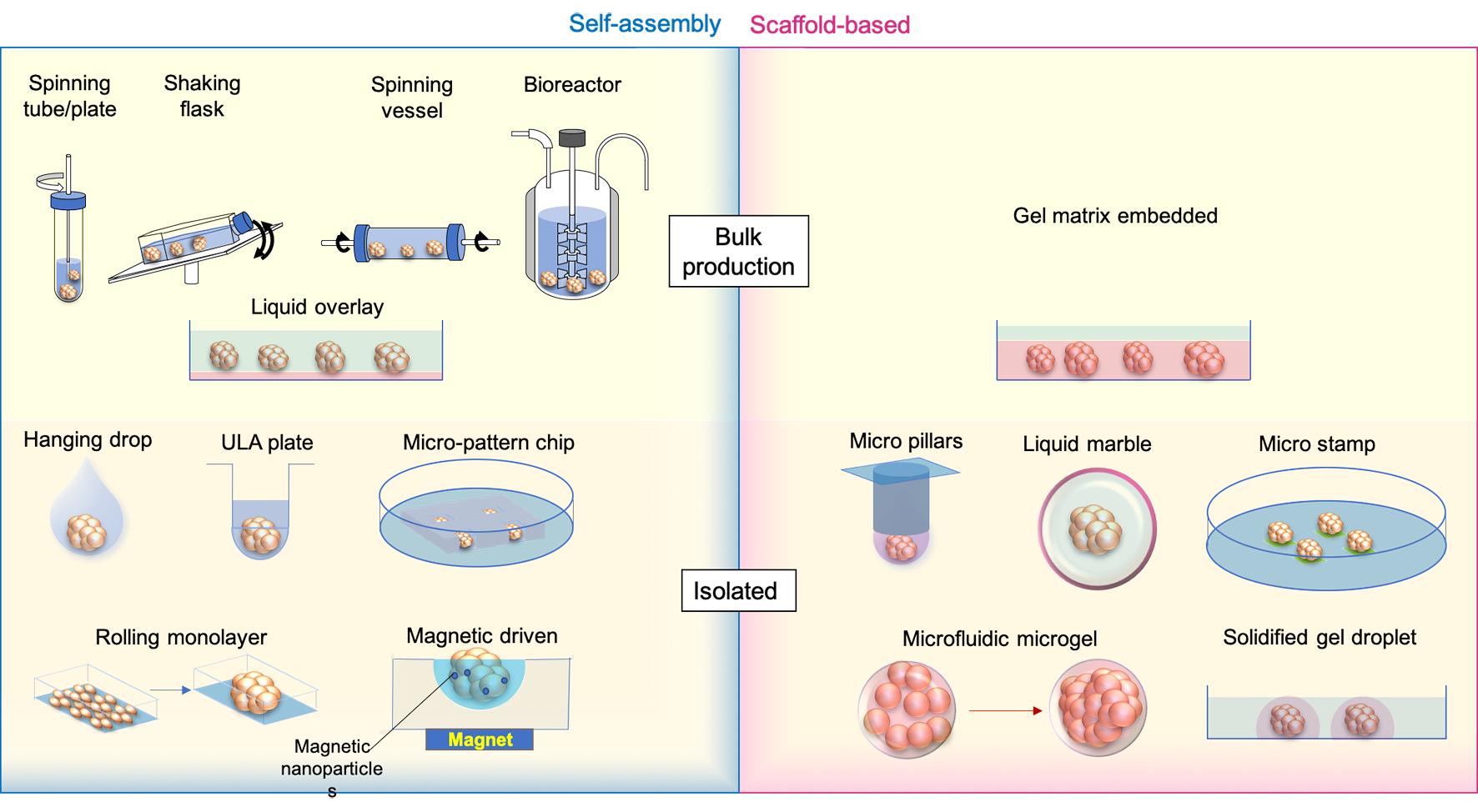
Figure 1. ‘Spheroid’ and ‘Organoid’ generation methods. Medium is labelled transparent blue, while scaffold gels are shown as transparent red. Micro stamps are presented as thin layers of green pads.
3.3.Self-assembly Cultures
In self-assembly spheroids/organoids, cells are compacted together by mechanical force, gravity, and the natural cohesive tendency of cells without pre-shaped scaffolds like hydrogels. The currently available methods include: (1) bulk organoid/spheroid production with spinning or shaking containers/bioreactors and culturing on gel-coated low-attachment dishes; and (2) isolated organoid generation with hanging drops, 96 or 384 low-attachment wells, micro-patterned plates, rolling monolayer, magnetic guiding clumps, etc.
To address the aims of drug screening or tests of therapeutic methods, making organoids or spheroids in batches is required. In forming a large number of embryoid bodies (EBs) from hiPSCs, spinner flasks or stirred-suspension bioreactors is a common method of producing and passaging EBs [22], following which the cells can be further differentiated into desired cell lineages, like neural, hepatic and cardiac cells [23–25]. This method has been used for making tumour spheroids from cell lines, such as HepG2 [26] and Human breast carcinoma cells (T-47D) [27]. Other strategies using the same approach in promoting the formation and maintenance of organoids or spheroids are shaking or rotating containers [28,29]. To reduce the requirements of costly equipment and large volume medium consumption in traditional spinning bioreactors, miniaturised bioreactors fitting standard 12 well plates have been developed and successfully applied to generate brain organoids which were tested for Zika virus (ZIKV) exposure [30]. Cells in these bioreactors or tubes/flasks/vessels/plates are promoted to aggregate by spinning or stirring power to avoid cell attachments to walls or settling to the bottom and increase cell-cell collisions [31]. These methods are productive and can be applied in long-term organoid or spheroid maintenance and drug exposures. However, the bioreactor system settings are relatively expensive; the produced organoids/spheroids are usually variable in size [32], and the method is not applicable to generate co-cultured organoids of mixed cell origins.
An economical way of generating spheroids or organoids in bulk is the liquid overlay method. To promote aggregation, cells are seeded on very low or none attachment Petri dishes, which are pre-coated with agarose [33], N-hexanoyl glycol chitosan (HGC) [34], PolyHema [35], and chitin gel [36], etc. The coating gels reduce cell adherence to the device but increase the cell-cell contacts, therefore, enhancing the formation of spheroids. Like spinning methods, the bulk production of organoids or spheroids with low attachment dishes is easy to handle and productive. Still, spheroids and organoids are highly heterogeneous in size, morphology, proliferation and reactivity to stimulations [37]. Due to the restrictions of static culture and limited oxygen and nutrition exchange rate, necrosis easily happens in large spheroids [38].
By contrast, other commonly applied self-assembly methods, like hanging drops, ultra-low attachment (ULA) wells and micro-patterning, are designed to produce isolated organoids or spheroids with controlled cell numbers and compositions.
Subsequent to the invention of the hanging-drop methodology in the 1880s, Harrison adapted it to constitute the first technique of generating 3D structures [39]. Cells inside hanging drops are prevented from direct interaction with the device and supporting substrates, forcing the cells to aggregate with each other [40]. The hanging-drop technique is widely applied in spheroid and organoid generation due to its simple method of handling, without the requirement of additional equipment, and its applicability of making co-cultivation spheroids/organoids with different cell populations [41]. To increase the productivity and standardisation of the method, new devices like hanging-drop plates [42], microfluidic channels [43] and pressure-controlled air chambers [44] for better loading of hanging drops have been recently invented and applied.
The ultra-low attachment (ULA) plate is a spheroid plate with a chemical coating surface minimizing protein and cell attachment to wells. It is an upgraded multiple-well version (96 or 384 wells) of the liquid overlay method in producing a single spheroid/organoid and is commercially available [3,45]. Rather than ready-to-use commercial plates, many studies developed their own ULA 96 well plates coated with various hydrophilic polymer surfaces such as agar or agarose [46,47] and hydrolysed poly(vinyl alcohol) [48].
Micro-patterned wells or chips, printed by micro-well or micro-chamber moulds using hydrogel, are substitutes for ULA plates. Unlike the ULA plate, micro-patterned wells or chambers are located as a rack at the bottom of a dish/well; cells are added to the dish and then aggregate at the bottom of each microchambers [49–51]. This approach scales up the number of spheroids or organoids produced and can provide the same exposure to testing medium or chemicals, making maintenance easier for anticancer drug screenings. The microwell spheroid chip has also been combined with a direct DNA analysis assay for genotoxicity screening to detect infrequent outcomes [52].
All these three self-assembly methods enable the co-culture of different cell types into isolated organoids/spheroids, increasing the homogeneity of spheroid/organoid morphology, the complexity of spheroid/organoid compositions and functions, and applicability to high throughput drug screening or toxicity test with reduced variance. Magnetic nanoparticles have also been introduced in combination with hanging drops, ULA plates, and micro-moulds for guiding the formation of the spheroids/organoids [53–55]. The external magnetic power improves the control of sphericity and reduces the duration of spheroid/organoid formation [55,56]. The drawback of these methods is that they are not very efficient in changing maintenance medium and exposure to screening ingredients. A lot of handling efforts are required at the generation, maintenance and screening stages.
In addition to the above methods, another productive way of generating spheroid/organoids with similar diameters is by rolling up monolayer cell cultures via compartmentation of cell culture surfaces utilising laser engraving (grid plates) [57]. Cells are cultured as monolayers inside the dish, subdivided by compartments; the plate then triggers the retraction at the edge of each compartment and therefore rolls the monolayer up as a spheroid [58]. Typically, about a hundred spheroids/ organoids can be made from one 60 mm dish. Not like the direct force of forming cell pellets, the rolling-up method takes a longer duration from the cell seeding as a monolayer to the final formation of spheroids, usually taking at least two weeks. The circularity of rolling-up spheroids is not as good as the ones derived from directly compacted cell aggregates [57].
3.4.Scaffold-Based Cultures
In relevance to the scaffold-free bulk and isolated spheroid/organoid generation methods, spheroids and organoids can also be formed and grown in a scaffold-based manner. The bulk production of spheroids within the ECM matrix is mainly used to make tumour spheroids from dissociated patient-derived tissues (surgical biolysis), from which the tumour cells proliferate and grow into spheroids at the embedded location of tissue cores [59]. The formed spheroids can then be processed for further expansion or drug screening.
Scaffold-based isolated spheroid/organoid generation is a significant improvement upon the hanging drop methods. Instead of providing the liquid drops for cell aggregation, cells are ‘fixed’ inside the gel drops attached to the bottom of inverted micropillars [60]. Pillars with spheroids/organoids are incubated by immersing the micropillars into maintenance medium cassettes, which are easily replaced for medium exchanges. The other way of culturing scaffold-based spheroids/organoids is encapsulating cells or cell-matrix mixture into a droplet. ‘Liquid marble’ is an example of the solid-phase spheroid formation method, in which the liquid with cells is enclosed and coated by hydrophobic powder [61]. Cells inside the ‘liquid marbles’ freely interact with each other and aggregate as uniformed spheroids/organoids within 24 hours [62]. Using the microfluidic system, a cell-hydrogel mixture can be encapsulated into microgels, where the cells proliferate and condense into spheroids/organoids [63,64]. The spheroids-in-gel can also be formed from cell pellets within the solidified matrigel droplets on the plate bottom, within which cell cores grow into spheroid/organoid inside each droplet [65]. On the other hand, spheroid-on-gel, which grows gradually from cells attached to printed micro-patterning hydrogel stamps, will finally form a single spheroid at each stamped area [5].
The scaffold of ECM will better supply the nutrients to cell growth and restrict the movement of cells from random floating. Whereas scaffolds may influence cell viability, distributions and interactive microenvironments, the sizes of scaffold-based isolated spheroid/organoid formation will be limited by the scaffold edges.
4.Preclinical Applications of Organoids
With the aim to produce in vitro systems similar to human organs, different protocols for generating human organoids have gradually developed in recent years. These protocols include hiPSC-differentiated organoids, and adult-stem cell (AdSC) derived organoids [ 66 ]. Here we summarise the launch times of human self-organising organoid models for major in vivo organ systems ( Figure 2 ). Among these models, we will introduce the preclinical applications of currently well-established intestinal, liver, pancreas, brain, and heart organoids in modelling development and disease, personalised medicine, drug discovery and screening.
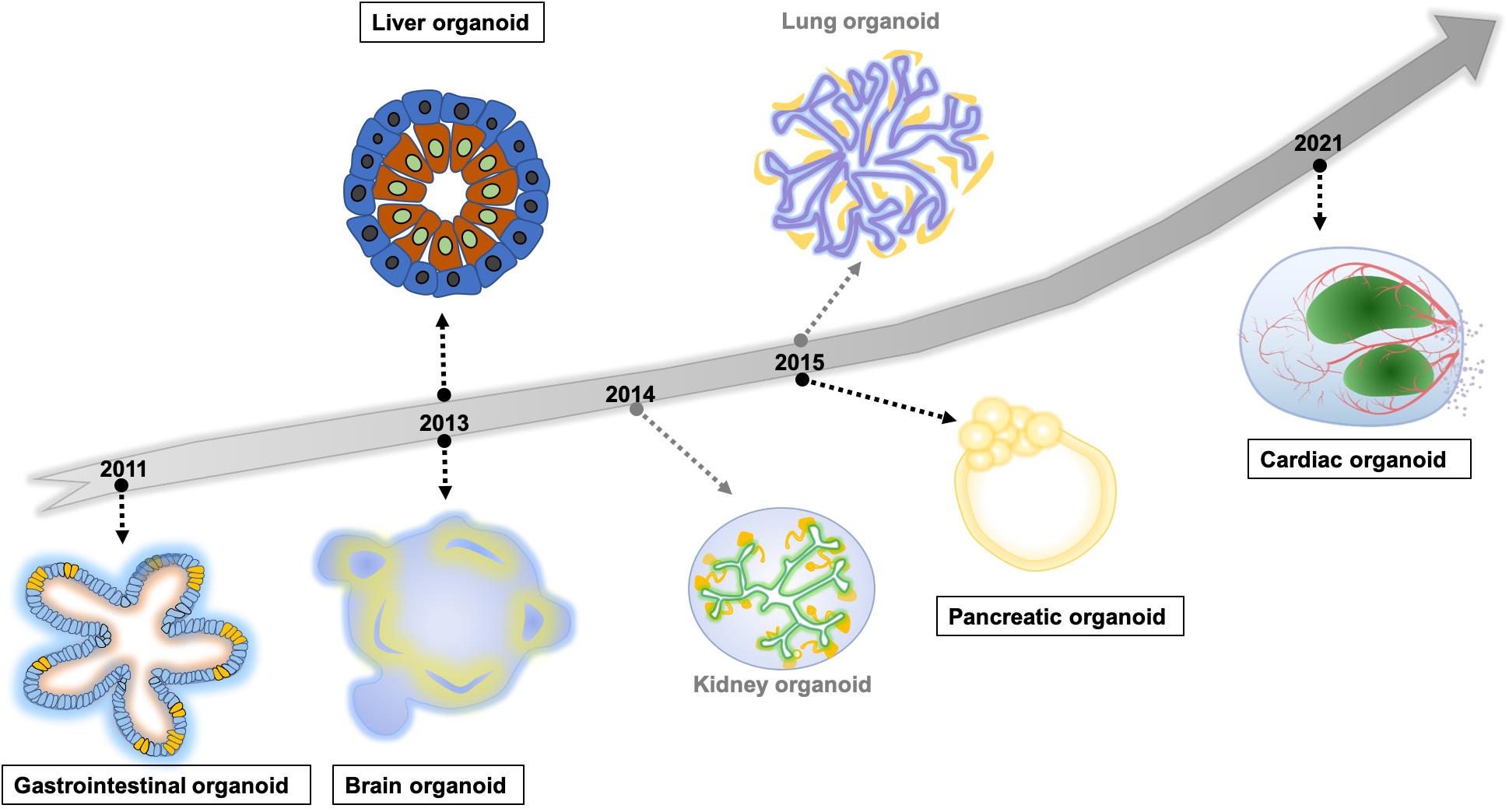
Figure 2. The timeline of hiPSC/hESC derived self-organising human organoids modelling major organs. The highlighted organoid models in rectangles are introduced in the section ‘Preclinical applications of organoids’.
4.1.Intestinal Organoids
The first human intestine organoid (IO) model was established from Lgr5(+) stem cells of small-intestinal crypts in 2009 [67]. Multiple crypts and villus-like epithelial domains are developed through self-organizing under long-time culture. Following the introduction of hiPSC techniques [68], iPSC-differentiated 3D intestinal organoids modelling different parts of intestinal systems and diseases were developed in 2011 [69]. A typical intestinal organoid recaptures the structures of polarized columnar epithelial cells, which pattern into distinct crypt and villus domains, containing different intestinal cell populations, such as enterocytes, goblet, and Paneth cells [70].
Besides the macroscopic morphology, intestinal organoids also recapture the functions, pharmacological dynamics, molecular expressions, and, to a degree, inter-individual heterogeneities. IOs generated from iPSCs with familial adenomatous polyposis (FAP-iPSCs) origins recapture the in vivo conditions of enhanced WNT activities and higher epithelial proliferation, which can be rescued by geneticin with its translational read-through activity [71]. Regardless of the cell sources from hiPSCs or patient tissue biobank, ‘Mini-gut’ IOs have been applied to modelling multiple intestinal diseases, including inflammatory bowel diseases, infectious diseases, and tumours [72–75] in addition to inherited syndromes. Pharmacokinetic and metabolic analyses of purified epithelial cells from iPSC-intestinal organoids indicate barrier functions and metabolizing enzyme activities mimicking human adult intestinal tissues [76]. Combined with sequencing technologies, patient-derived intestinal organoids from colorectal and gastroesophageal cancers show high agreements with original tumour tissues in phenotypes and genotypes, reflecting the heterogeneities between human individuals [77]. Targeted organoid sequencing (TORNADO-seq), adapting RNA sequencing with organoid drug screenings, not only provides an in-depth molecular understanding of active drug motions but also distinguishes differentiation-inducing drugs, a common mechanism for efficacious drugs targeting against cancer organoids [78]. The well-matched morphological and molecular properties between IOs and intestinal tissues endorse IOs’ critical roles in discovering, screening, and evaluating novel antibiotics, regeneration medicine, anticancer drugs, and personalised therapies.
4.2.Liver Organoids
Depending on the liver’s capability of regenerating itself [79], liver organoids were developed from primary LGR5+ biliary cells of injured mouse liver in 2013 [80] and from patients’ liver samples with α-1 antitrypsin (A1AT) deficiency and Alagille syndrome (ALGS) in 2015 [81]. The first iPSC-derived human liver organoid model (iPSC-derived liver bud (iPSC-LB)) was developed by co-culturing iPSC-derived endoderm (iPSC-HE) with human umbilical vein endothelial cells (HUVECs) and human mesenchymal stem cells (MSCs) in 2013 [82]. Later, human hepatic organoids were directly differentiated from iPSCs using sequential stimulations of growth factor and chemicals, and a model constructed by reassembling human foetal liver progenitor cells was developed. These can modulate human liver embryonic developments, hepatobiliary organogenesis, complex liver structures of sheeting hepatocytes and bile duct-like lumens with a surrounding epithelial layer of cholangiocytes [83,84]. Profiling with scRNAseq distinguishes the variety of hepatocytic zonal cell populations inside the hiPSC-derived liver organoids, including immature and adult-like hepatocytes [85]. Additional to the phenotypic similarities in cell populations and structures, liver organoids retain functions of producing serum proteins, detoxifying and metabolizing drugs, and responding to regenerative and inflammatory stimulations; which in turn provides liver organoids with the capability to recapture the toxic responses to medications with known hepatotoxicity [86] and the applicability to high throughput toxicity screenings [87].
Liver organoids have been applied to model different hepatic diseases. On introducing the mutation of C829X in JAG1, liver organoids can display an Alagille syndrome (ALGS)-like phenotype [83]. Organoids derived from patients with defective CFTR (Cystic Fibrosis Transmembrane Conductance Regulator) genes emulate cystic fibrosis (CF) in reactivity to increased cAMP levels [88]. Liver organoids modelling viral hepatitis, by infecting with Hepatitis B (HBV) [89] or Hepatitis C (HCV) [90], are capable of being tested on anti-HBV/HCV activities and drug-induced hepatic toxicity. Combined with transcriptomic analysis, patient-derived liver organoids exhibit early cancer gene signatures similar to hepatocellular carcinoma [91]. The similarity of patient-derived organoids at the molecular level suggests their preserved heterogeneity in responding to drugs and chemotherapy [92,93]. Due to the limitation that patients-derived liver cancer organoids need to be sampled at late stages when the tumour is poorly differentiated with high proliferation rates [94], CRISPR/Cas9 technology has been applied to model liver cancers by generating mutations in healthy liver organoids [95]. Both healthy and cancer liver organoids are of high sensitivity and specificity (about 90%) in screening anticancer compounds and the cholestatic and mitochondrial toxicity in marketed drugs [94,96,97].
4.3.Pancreatic Organoids
The early protocols generated pancreatic spheres from pancreatic ductal and acinar progenitor cells obtained from primary human pancreas progenitors or monolayer differentiated hESCs [98]. These methods formed hollow spheroids without elongating ducts, rather than organoids [99,100]. These spheroids preserve proliferation properties, allowing serial passages and maintaining progenitor markers SOX9/PDX1/NKX6.1 [101]. True pancreatic organoids were established from hiPSCs and patient-derived pancreas progenitors (tumour) in 2015 [98]. Endoderm-derived or hiPSC differentiated progenitors further proliferated and reorganised into polarised exocrine organoids with ductal and acinar-like structures, displaying appropriate gene expression patterns and functional hallmarks similar to the human pancreas [98,102]. Other studies improved the pancreatic organoid complexities by branching ducts with short-range growth inhibitory signals [103] or segregating endocrine progenitor cells and acinar cells to either the centre or tube tips with high levels of FGF10 [100].
Pancreatic ductal adenocarcinoma (PDAC), as the third leading cause of cancer-related mortality, is highly aggressive [103]. Therefore, pancreatic cancers become the primary disease modelling purpose for generating pancreatic organoids. Living biobanks of patient-derived pancreatic cancer organoid lines were established and applied to high-throughput drug screening [104,105]. Similar to other patient-derived organoids, human PDAC organoids maintain their original heterogeneity as the primary PDAC tissues [106].
With a joint strategy incorporating 3D bioprinting technology, pancreatic cancer organoids have been applied to a pilot cytotoxicity screen of ~3300 approved drugs, from which stable cytotoxic reagents across different types of pancreatic cancers were identified [107]. Together with CRISPR-Cas9, pancreatic organoids are applied to detect drug-gene interactions, whereby ARID1A mutations were found to be associated with increased sensitivity to the kinase inhibitors, like dasatinib and VE-821 [106]. Omics data on 84 pancreatic cancer organoid lines indicate the association between chromatin accessibility signatures and drug sensitivity based on analysis of 283 epigenetic-related chemicals and five chemotherapeutic drugs [108], providing deep sights into drug-tissue reactive mechanisms.
As a critical metabolic organ, the endocrine cells of the pancreas regulate circulating glucose levels via insulin and glucagon. Pancreatic islets-on-chip and combined two-organ chip system of islet and liver have also been developed and applied to model non-cancer diseases of cystic fibrosis (CF) and type 2 diabetes for evaluation on glucose tolerance and effects of drug treatment [109].
4.4.Brain Organoids
The evolutionary uniqueness of the human brain, and the length of the human lifespan, make many brain diseases difficult to study via model organisms. In vitro models of the human brain are, therefore, critical for modelling brain development and diseases. In 2013, a human cerebral organoid was developed from hiPSCs and applied to model microcephaly with RNA interference [110]. Progenitor zone organisation, cellular layers, neurogenesis, and gene expressions are featured in these organoids [30]. Progressively, strategies to improve the culture conditions and constructions are gradually being introduced, such as culturing inside the human brain extracellular matrix (BME) and using a microfluidic device to reduce organoids’ variability. Proofs show that BME can increase the radial glial cell population, volumetric augmentation, and promote cortical layer development and electrophysiological functions in brain organoids [111]. Gradually, brain organoids emulated different regions of human brains, including the neocortex, rostral and caudal cortices, cortical hem and choroid plexus, dorsal cortex, midbrain, hypothalamus and hippocampus [30,112]. Furthermore, there are studies trying to fuse the different parts of the brain organoids to increase their complexity. Human forebrain organoids (hFOs) and human midbrain organoids (hMOs) have been combined with the controllable acoustofluidic alignment into assembloids to study the neuron projection regulations, maturation, and neural progenitor cell (NPC) division inside the bud [113,114].
Regardless of the limitations of current organoids, including incomplete cell type diversity, slow maturation and low reproducibility [114], these models have been widely applied to investigate human brain disorders and cancers: developmental disorders like microcephaly, epilepsy, and autism spectrum disorder (ASD); neurodegenerative diseases like Niemann-Pick disease, Parkinson’s disease (PD), Alzheimer’s disease (AD), Huntington’s disease (HD); and brain cancers including glioblastoma multiforme (GBM) [115]. Zika virus (ZIKV) infection causes microcephaly [116]; brain organoids modelling the ZIKV-induced microcephaly show premature neural progenitor differentiation together with centrosome perturbation, leading to impaired neurogenesis and thinned cortical layer [117]. Niemann-Pick disease type C (NPC), a neurodegenerative lysosomal storage disorder, is recaptured by the NPC iPSC-derived organoids conceiving NPC1 or NPC2 mutations. These NPC brain organoids are smaller in size with accumulated cholesterol and impaired neuronal differentiation, similar to the major phenotypes of NPC patients and reversible properties by know drugs of valproic acid and HPBCD [118].
Patient-derived glioma cerebral organoids recapitulate the histological characteristics, chemotherapeutic drug responses, and clinical progression of GBM [119]. Assays using brain organoids specify glioma stem cells (GSCs)’s activities in invasion, integration, interaction with mature neurons, and reactivity to pharmacological agents; for example, the inhibitor of ADAM10, GI254023X, can prevent the integration of GSCs into the organoids [120].
In addition, human brain organoids have been investigated for future regenerative medicine by transplanting into an adult mouse brain disorder model [121]. Microdevices like microfluidic and organoid-on-a-chip technologies help improve the precise culture maintenance and fitness to real-time monitoring of organoid development processes and pharmaceutical reactions [122]. Adapting the CRISPR-Cas9 technology, organoids with gain or loss of function mutations can be generated for observations of early-stage development and neurodegeneration [123]. Benefiting from high-content screening (HCS), multiple electrode array (MEA) and transcriptome data, high-throughput screening of blood-brain barrier-permeable drugs can be analysed with mathematical modelling to figure out the molecular pathways and relevant genetic factors associated with the external application of drugs [124–126]. The phenotypic, electronic, and molecular data have led to a better understanding of neurodevelopmental disorders with complex genetic and environmental etiologies [127], as well as drug effects together with receptor functions and synaptic plasticity [124]. With the multiple developed strategies in improving and specifying brain organoid generation, qualification and evaluation, brain organoids are becoming more and more consistent in drug responses, predisposing to large-scale screening and discovering new drugs and promoting precision medicine therapies.
4.5.Cardiac Organoids
As the first functional organ in human embryonic development, the complex cell populations and highly hierarchical structures of the heart make it challenging to develop in vitro 3D models. There are diverse cardiac cell populations inside a heart, such as sinoatrial nodal cells, atrioventricular cells, atrial cells, ventricular cells, and Purkinje cells [128]. The majority of the currently reported ‘cardiac organoids’ are formed by a forced combination of primary or hiPSC-CM together with endothelial, fibroblast or stromal cells. Accurately, these combined cell clusters with simple cell distributions should be named ‘cardiac spheroid’. These spheroids resemble the in vivo cardiac tissue better with similar cell components, expression patterns and stable contractility than 2D hiPSC-CMs [129]. Using patient-derived cardiomyocytes, the ‘cardiac spheroid’ has been applied to model the heart tissues of genetic disorders, such as cardiomyopathy [130]. They are typically condensed in texture with evenly distributed cardiomyocytes with or without other cell types, recapturing the beating activities, cell properties and responses to external stimulation, including drugs. Also, ‘cardiac spheroid’, as a cluster of aggregated cells and matrix, incorporates the oxygen-diffusion gradient, yielding the potential for modelling myocardial infarctions by mimicking the infarcted areas, and associated changes in pathological metabolism, fibrosis and calcium handling [131]. These co-cultured ‘cardiac spheroids’ are also applicable for cardiotoxicity or developmental toxicity screening [132]. By quantifying the contraction velocity, beating rates and durations, ‘cardiac spheroids’ can be applied to evaluate exposures to drugs or environmental toxins [133], whereby the known teratogens can impair ‘cardiac spheroid’ organization [134]. Heart-Dyno is a device of 96-well plate with two casted polydimethylsiloxane (PDMS) inserts in each well, surrounding which the condensed hPSC-CMs and stromal cells will form bundle-like cardiac spheroids. This platform can be applied to the high-throughput functional screening of metabolic substrates and chemicals for potential regenerative medicine [135]. In combination with 3D bioprinting, human engineered heart tissue (hEHT), also referred to as a ‘cardiac organoid’ in many studies, is fabricated as endothelialized myocardium for modelling mature adult heart tissues, which are applied as a platform for evaluating cardiovascular toxicity [136].
However, the above co-cultured ’cardiac spheroid’ or fabricated ‘engineered heart tissue’ are not self-organised cardiac organoids [137]. A self-organising vascularised organoid with sprouting capillary network was reported in 2019 [138], while the first report of self-organizing cardiac organoids emulating cardiogenesis was published in 2021 [139]. These cardiac organoids contain divided layers of myocardial cells, endocardial-like cells, distinct anterior/posterior foregut endoderm tissues and a vascular network [140,141]. Unlike the traditional free-floating embryoid body (EB) differentiation into high purity of cardiomyocytes, Drakhlis’s method of differentiating Matrigel embedded EBs of defined size (5000 cells/EB) via similar biphasic WNT pathway modulation gives rise to a complex and highly structured cardiac organoid with separated layers [140]. Rossi et al. achieved their cardiac organoid (called gastruloids) with mouse ECSs, recapturing the early cardiogenesis with the separated spatial distribution of first heart field (FHF) and second heart field (SHF) progenitor cells and tube-like structures [130]. At the same time, Lewis-Israeli et al. developed a remarkable cardiac organoid (inducted by three-step Wnt signalling modulation, including the first WNT activation, WNT inhibition and a second WNT activation exposure on day 7) with cavity formation, containing major heart cell types of cardiomyocytes, endocardial cells, and epicardial cells [142]. Among the current methods of generating self-organised cardiac organoids, with either single or multiple chamber-like cavities [143], separated atrium and ventricle zones [144], or first and second heart fields like regions [145], the three-step Wnt modulation strategy is relatively easy to handle and more frequently applied to modelling cardiac development and diseases. Starting from the same hESC or hiPSC line, cardiac organoids are differentiated under healthy and diabetic conditions (high insulin and glucose), simulating the pregestational diabetes condition for evaluation of its influence on heart development [146]. Also, cardiac organoids have been applied to model complex metabolic disorders, as well as congenital heart defects [142]. Due to the newest and varied nature of the current available self-organising cardiac organoid generation methods, there are not yet many studies that conduct high-throughput drug screening with self-organising cardiac organoids.
5.Future Directions
With respect to structures, spheroids are relatively simpler than organoids. Compared to spheroids, organoids are more complex in tissue fabrication and cell compositions, retaining self-reorganising properties. However, looking at culture methods and cell compositions, there is no definitive difference between spheroids, organoids, and other nomenclatures like microtissues. Especially in the functional evaluation and drug screening studies, the notion of ‘organoid’ is preferred in use, regardless of their generation methods. Therefore, a quantifiable standard for separating spheroids and organoids, as well as other similar definitions, such as engineered tissues, microtissues, mini-organs, etc., could be of use in the field.
Although the organoids recapture the organ structures, properties and functions well, the self-assembly and self-reorganising characteristics lead to marked heterogeneity in sizes and shapes, which could affect the interpretation of high-throughput screening experiments. Various applied scaffolding matrices may also affect repsonses. Controlling loading devices like microfluidic systems promotes the uniform phenotypes of organoids; further development of these systems and their applications remain necessary.
Looking forward, a newly developed ‘body-on-a-chip' system, integrates multi-organoids modelling liver, cardiac, lung, vascular, testis, colon, and brain on one platform, which is applicable to study the interdependent metabolism and drug effects across different tissue types [147]. The organ-on-chip system, with engineered separated chambers for loading different organoids, provides a more-controllable and conducive environment for learning the interaction and communications between co-cultured ‘mini organs’ [148]. Multi-Organs-on-Chip (MOoC) combining the liver organoids and ‘cardiac organoids’ is a remarkable example of learning the drug-related effects and predicting liver metabolism on off-target organs during preclinical testing, which will be meaningful to improve drug safety screening prior to clinical uses [149]. Simultaneously, tumour-on-a-chip with combined components of tumour, stromal and immune cells, providing the microenvironment for cancer-immune system crosstalk over the vascularised endothelial barrier, offers a potential screening platform for immunotherapies [150]. As a direction for future evolutions of preclinical screening and investigations of drugs, multiple organ-on-chip systems resemble the human in vivo environment better and will likely prove indispensable in looking at inter-tissue communications, side effects and off-target toxicity.
Supplementary Materials: The following supporting information can be downloaded at: https://www. sciltp. com/ journals/ijddp/2022/1/188/56
Author Contributions: YL and HX complete the literature searches, review of included studies, writing the first version of the manuscript. SA, XW, and BK contributed to the manuscript with their thoughtful inputs on the review structures and contents. All authors have read and agreed on the manuscript for submission.
Funding: YL is supported by BHF Programme Grant RG/F/21/110050 and RG/15/12/31616. BK holds a British Heart Foundation Personal Chair and the BHF Accelerator Awards AA/18/4/34221. HX and XW are supported by BHF grants PG/17/31/32988 and PG/19/53/34499.
Data Availability Statement: Not applicable.
Acknowledgments: Thanks to the contribution of Siew Hon Teay for the artwork of figures.
Conflict of Interest: The authors declare no conflict of interest.
References
- Jaroch K.; Jaroch A.; Bojko B. Cell cultures in drug discovery and development: The need of reliable in vitro-in vivo extrapolation for pharmacodynamics and pharmacokinetics assessment. J. Pharm. Biomed. Anal., 2018, 147:297-312, doi: 10.1016/j.jpba.2017.07.023. DOI: https://doi.org/10.1016/j.jpba.2017.07.023
- Imamura Y.; Mukohara T.; Shimono Y.; et al. Comparison of 2D- and 3D-culture models as drug-testing platforms in breast cancer. Oncol. Rep., 2015, 33(4):1837-43, doi: 10.3892/or.2015.3767. DOI: https://doi.org/10.3892/or.2015.3767
- Nowacka M.; Sterzynska K.; Andrzejewska M.; et al. Drug resistance evaluation in novel 3D in vitro model. Biomed. Pharmacother., 2021, 138:111536, doi: 10.1016/j.biopha.2021.111536. DOI: https://doi.org/10.1016/j.biopha.2021.111536
- Roper S.J.; Linke F.; Scotting PJ.; et al. 3D spheroid models of paediatric SHH medulloblastoma mimic tumour biology, drug response and metastatic dissemination. Sci. Rep., 2021, 11(1):4259, doi: 10.1038/s41598-021-83809-6. DOI: https://doi.org/10.1038/s41598-021-83809-6
- Li S.; Yang K.; Chen X.; et al. Simultaneous 2D and 3D cell culture array for multicellular geometry, drug discovery and tumor microenvironment reconstruction. Biofabrication, 2021, 13(4), doi: 10.1088/1758-5090/ac1ea8. DOI: https://doi.org/10.1088/1758-5090/ac1ea8
- Fontoura J.C.; Viezzer C.; dos Santos F.G.; et al. Comparison of 2D and 3D cell culture models for cell growth, gene expression and drug resistance. Mater. Sci. Eng. C. Mater. Biol. Appl., 2020,107:110264, doi:10.1016/j.msec.2019.110264. DOI: https://doi.org/10.1016/j.msec.2019.110264
- Edmondson R.; Broglie J.J.; Adcock A.F.; et al. Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors. Assay Drug Dev. Technol., 2014, 12(4):207-18, doi:10.1089/adt.2014.573. DOI: https://doi.org/10.1089/adt.2014.573
- Kapałczyńska M.; Kolenda T.; Przybyła W.; et al. 2D and 3D cell cultures - a comparison of different types of cancer cell cultures. Arch. Med. Sci., 2018,14(4):910-9, doi: 10.3390/ijms222212200. DOI: https://doi.org/10.3390/ijms222212200
- Wilson H. A new method by which sponges may be artificially reared. Science, 1907, 25(649):912-5, doi: 10.1126/science.25.649.912, doi: 10.1126/science.25.649.912. DOI: https://doi.org/10.1126/science.25.649.912
- Steinberg M.S. The problem of adhesive selectivity in cellular interactions. Cellular membranes in development. 22: Academic Press New York; 1964. p. 321-66. DOI: https://doi.org/10.1016/B978-0-12-395533-3.50015-6
- James A.W.; Jennifer J.; Swiergiel V.S.M.; et al. Embryonic Stem Cell Lines Derived from Human Blastocysts. Science, 1998, 282(5391):1145, doi: 10.1126/science.282.5391.1145. DOI: https://doi.org/10.1126/science.282.5391.1145
- Shannon J.M.; Mason R.J.; Jennings S.D. Functional differentiation of alveolar type II epithelial cells in vitro: effects of cell shape, cell-matrix interactions and cell-cell interactions. Biochim. Biophys. Acta., 1987, 931(2):143-156, doi:10.1016/0167-4889(87)90200-x. DOI: https://doi.org/10.1016/0167-4889(87)90200-X
- Pollard R.T.; Salter I.; Sanders R.J.; et al. Southern Ocean deep-water carbon export enhanced by natural iron fertilization. Nature, 2009, 457(7229):577-80, doi:10.1038/nature07716 . DOI: https://doi.org/10.1038/nature07716
- Corrò C.; Novellasdemunt L.; Li V.S.W. A brief history of organoids. Am. J. Physiol. Cell Physiol., 2020, 319(1):C151-C65, doi:10.1152/ajpcell.00120.2020. DOI: https://doi.org/10.1152/ajpcell.00120.2020
- Huch M.; Knoblich J.A.; Lutolf M.P.; et al. The hope and the hype of organoid research. Development. 2017,144(6):938-41, doi: 10.1242/dev.150201. DOI: https://doi.org/10.1242/dev.150201
- Xu H.; Jiao Y.; Qin S.; et al. Organoid technology in disease modelling, drug development, personalized treatment and regeneration medicine. Exp. Hematol. Oncol., 2018,7:30, doi: 10.1186/s40164-018-0122-9. DOI: https://doi.org/10.1186/s40164-018-0122-9
- Karolak A.; Poonja S.; Rejniak K.A. Morphophenotypic classification of tumor organoids as an indicator of drug exposure and penetration potential. PLOS Computational Biology. 2019,15(7):e1007214, doi: 10.1186/s40164-018-0122-9. DOI: https://doi.org/10.1371/journal.pcbi.1007214
- Gunti S.; Hoke A.T.K.; Vu K.P.; et al. Organoid and Spheroid Tumor Models: Techniques and Applications. Cancers (Basel). 2021,13(4), doi: 10.3390/cancers13040874. DOI: https://doi.org/10.3390/cancers13040874
- Sutherland R.M.; McCredie J.A.; Inch W.R. Growth of multicell spheroids in tissue culture as a model of nodular carcinomas. J. Natl. Cancer Inst., 1971,46(1):113-20.
- Sutherland R.M.; Inch W.R.; McCredie J.A.; et al. A multi-component radiation survival curve using an in vitro tumour model. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med., 1970,18(5):491-5. DOI: https://doi.org/10.1080/09553007014551401
- Sakalem M.E.; De Sibio M.T.; da Costa F.A.d.S.; et al. Historical evolution of spheroids and organoids, and possibilities of use in life sciences and medicine. J. Biotechnology. 2021,16(5):2000463, https://doi.org/10.1002/biot.202000463. DOI: https://doi.org/10.1002/biot.202000463
- Kurosawa H. Methods for inducing embryoid body formation: in vitro differentiation system of embryonic stem cells. J. Biosci. Bioeng., 2007;103(5):389-398. doi:10.1263/jbb.103.389. DOI: https://doi.org/10.1263/jbb.103.389
- Liyang G.; Abdullah S.; Rosli R; et al. Neural Commitment of Embryonic Stem Cells through the Formation of Embryoid Bodies (EBs). Malays. J. Med. Sci., 2014,21(5):8-16.
- Imamura T.; Cui L.; Teng R.; et al. Embryonic stem cell-derived embryoid bodies in three-dimensional culture system form hepatocyte-like cells in vitro and in vivo. Tissue. Eng., 2004,10(11-12):1716-24, doi: 10.1089/ten.2004.10.1716. DOI: https://doi.org/10.1089/ten.2004.10.1716
- Zhang J.; Wilson G.F.; Soerens A.G.; et al. Functional cardiomyocytes derived from human induced pluripotent stem cells. Circ. Res., 2009,104(4):e30-41, doi: 10.1161/CIRCRESAHA.108.192237. DOI: https://doi.org/10.1161/CIRCRESAHA.108.192237
- Castañeda F.; Kinne R .K. Short exposure to millimolar concentrations of ethanol induces apoptotic cell death in multicellular HepG2 spheroids. J. Cancer. Res. Clin. Oncol., 2000,126(6):305-10, doi: 10.1007/s004320050348. DOI: https://doi.org/10.1007/s004320050348
- Bartholomä P.; Impidjati.; R.-M.A.; Zhang, Z.; et al. A More Aggressive Breast Cancer Spheroid Model Coupled to an Electronic Capillary Sensor System for a High-Content Screening of Cytotoxic Agents in Cancer Therapy: 3-Dimensional In Vitro Tumor Spheroids as a Screening Model. J. Biomol. Screen. 2005,10(7):705-14, doi: 10.1177/1087057105277841. DOI: https://doi.org/10.1177/1087057105277841
- Niibe K.; Ohori-Morita Y.; Zhang M.; et al. A Shaking-Culture Method for Generating Bone Marrow Derived Mesenchymal Stromal/Stem Cell-Spheroids With Enhanced Multipotency in vitro. Front. Bioeng. Biotechnol., 2020; 8:590332, doi:10.3389/fbioe.2020.59033. DOI: https://doi.org/10.3389/fbioe.2020.590332
- Carpenedo R.L.; Sargent C.Y.; McDevitt T.C. Rotary suspension culture enhances the efficiency, yield, and homogeneity of embryoid body differentiation. Stem. Cells. 2007,25(9):2224-34, doi:10.1634/stemcells.2006-0523. DOI: https://doi.org/10.1634/stemcells.2006-0523
- Qian X.; Nguyen H.N.; Song M.; et al. Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure. Cell. 2016,165(5):1238-54, doi:10.1016/j.cell.2016.04.032 . DOI: https://doi.org/10.1016/j.cell.2016.04.032
- Achilli T.M., Meyer J.; Morgan J.R. Advances in the formation, use and understanding of multi-cellular spheroids. Expert Opin. Biol. Ther., 2012,12(10):1347-60, doi:10.1517/14712598.2012.707181. DOI: https://doi.org/10.1517/14712598.2012.707181
- Velasco V.; Shariati S.A.; Esfandyarpour R. Microtechnology-based methods for organoid models. Microsyst. Nanoeng., 2020,6(1):76, doi:10.1038/s41378-020-00185-3. DOI: https://doi.org/10.1038/s41378-020-00185-3
- Li M.; Fu T.; Yang S.; et al. Agarose-based spheroid culture enhanced stemness and promoted odontogenic differentiation potential of human dental follicle cells in vitro. In Vitro Cell Dev. Biol. Anim., 2021,57(6):620-630. doi:10.1007/s11626-021-00591-5. DOI: https://doi.org/10.1007/s11626-021-00591-5
- Cho M.O.; Li Z.; Shim H-E.; et al. Bioinspired tuning of glycol chitosan for 3D cell culture. NPG Asia Mater., 2016,8(9):e309-e, https://doi.org/10.1038/am.2016.130. DOI: https://doi.org/10.1038/am.2016.130
- Rasouli R.; Tabrizian M. Rapid Formation of Multicellular Spheroids in Boundary-Driven Acoustic Microstreams. Small. 2021,17(39):2101931, https://doi.org/10.1002/smll.202101931. DOI: https://doi.org/10.1002/smll.202101931
- Inubushi Y.; Tachibana A. Uniform spheroid formation on a laboratory-made, low cell attachment surface consisting of a chitin sheet. Biosci. Biotechnol. Biochem., 2020,84(5):997-1000, doi:10.1080/09168451.2020.1714423. DOI: https://doi.org/10.1080/09168451.2020.1714423
- Carlsson J.; Yuhas J.M. Liquid-overlay culture of cellular spheroids. Recent Results Cancer Res., 1984,95:1-23. DOI: https://doi.org/10.1007/978-3-642-82340-4_1
- Mehta G.; Hsiao A.Y.; Ingram M.; et al. Opportunities and challenges for use of tumor spheroids as models to test drug delivery and efficacy. J. Control Release. 2012;164(2):192-204. doi:10.1016/j.jconrel.2012.04.045. DOI: https://doi.org/10.1016/j.jconrel.2012.04.045
- Alhaque S.; Themis M.; Rashidi H . Three-dimensional cell culture: from evolution to revolution. Philos. Trans. R. Soc. Lond. B Biol. Sci., 2018,373(1750):20170216. doi:10.1098/rstb.2017.0216. DOI: https://doi.org/10.1098/rstb.2017.0216
- Timmins N.E.; Nielsen L.K. Generation of Multicellular Tumor Spheroids by the Hanging-Drop Method. In: Hauser H, Fussenegger M, editors. Tissue Engineering. Totowa, NJ: Humana Press; 2007. p. 141-51. DOI: https://doi.org/10.1007/978-1-59745-443-8_8
- Foty R. A simple hanging drop cell culture protocol for generation of 3D spheroids. J. Vis. Exp., 2011,(51), doi:10.3791/2720. DOI: https://doi.org/10.3791/2720-v
- Jeong Y.; Tin A.; Irudayaraj J. Flipped Well-Plate Hanging-Drop Technique for Growing Three-Dimensional Tumors. Front. Bioeng. Biotechnol., 2022,10:898699. doi:10.3389/fbioe.2022.898699. DOI: https://doi.org/10.3389/fbioe.2022.898699
- Rodoplu D.; Matahum J.S.; Hsu C.-H. A microfluidic hanging drop-based spheroid co-culture platform for probing tumor angiogenesis. Lab. Chip. 2022,22(7):1275-85, doi:10.1039/d1lc01177d. DOI: https://doi.org/10.1039/D1LC01177D
- Cho C.-Y.; Chiang T.-H.; Hsieh L.-H.; et al. Development of a Novel Hanging Drop Platform for Engineering Controllable 3D Microenvironments. Front. Cell Dev. Biol., 2020,8. doi:10.3389/fcell.2020.00327. DOI: https://doi.org/10.3389/fcell.2020.00327
- Grandhi T.S.P.; To J.; Romero A.; et al. High-throughput CRISPR-mediated 3D enrichment platform for functional interrogation of chemotherapeutic resistance. Biotechnol. Bioeng., 2021,118(8):3187-99, doi:10.1002/bit.27844. DOI: https://doi.org/10.1002/bit.27844
- Vinci M.; Gowan S.; Boxall F.; et al. Advances in establishment and analysis of three-dimensional tumor spheroid-based functional assays for target validation and drug evaluation. BMC Biol., 2012,10:29, doi:10.1186/1741-7007-10-29. DOI: https://doi.org/10.1186/1741-7007-10-29
- Liao W.; Wang J.; Xu J.; et al. High-throughput three-dimensional spheroid tumor model using a novel stamp-like tool. J. Tissue Eng., 2019, 10:2041731419889184, doi:10.1177/2041731419889184. DOI: https://doi.org/10.1177/2041731419889184
- Molyneaux K.; Wnek M.D.; Craig S.E.L.; et al. Physically-cross-linked poly(vinyl alcohol) cell culture plate coatings facilitate preservation of cell–cell interactions, spheroid formation, and stemness. J. Biomed. Mater. Res. B. Appl. Biomater.,. 2021, 109(11):1744-53, doi:10.1002/jbm.b.34832. DOI: https://doi.org/10.1002/jbm.b.34832
- Otsuka H.; Sasaki K.; Okimura S.; et al. Micropatterned co-culture of hepatocyte spheroids layered on non-parenchymal cells to understand heterotypic cellular interactions. Sci. Technol. Adv. Mater., 2013, 14(6):065003, doi:10.1088/1468-6996/14/6/065003. DOI: https://doi.org/10.1088/1468-6996/14/6/065003
- Valdoz J.C.; Jacobs D.J.; Cribbs C.G.; et al. An Improved Scalable Hydrogel Dish for Spheroid Culture. Life (Basel). 2021, 11(6), doi:10.3390/life11060517. DOI: https://doi.org/10.3390/life11060517
- Lee J.M; Park D.Y.; Yang L.;et al. Generation of uniform-sized multicellular tumor spheroids using hydrogel microwells for advanced drug screening. Sci. Rep., 2018, 8(1):17145, doi:10.1038/s41598-018-35216-7. DOI: https://doi.org/10.1038/s41598-018-35216-7
- Chao C.; Ngo L.P.; Engelward B.P. SpheroidChip: Patterned Agarose Microwell Compartments Harboring HepG2 Spheroids are Compatible with Genotoxicity Testing. ACS Biomater. Sci. Eng., 2020, 6(4):2427-39, doi:10.1021/acsbiomaterials.9b01951. DOI: https://doi.org/10.1021/acsbiomaterials.9b01951
- Ho V.H.B.; Müller K.H.; Barcza A.; et al. Generation and manipulation of magnetic multicellular spheroids. Biomaterials. 2010, 31(11):3095-102, doi:10.1016/j.biomaterials.2009.12.047. DOI: https://doi.org/10.1016/j.biomaterials.2009.12.047
- Gaitán-Salvatella I.; López-Villegas E.O.; González-Alva P.; et al. Case Report: Formation of 3D Osteoblast Spheroid Under Magnetic Levitation for Bone Tissue Engineering. Front. Mol. Biosci., 2021, 8, doi:10.3389/fmolb.2021.672518. DOI: https://doi.org/10.3389/fmolb.2021.672518
- Perez J.E.; Nagle I.; Wilhelm C. Magnetic molding of tumor spheroids: emerging model for cancer screening. Biofabrication. 2021, 13(1):015018, doi:10.1088/1758-5090/abc670. DOI: https://doi.org/10.1088/1758-5090/abc670
- Kim J.A. ; Choi J .-H.; Kim M.;et al. High-throughput generation of spheroids using magnetic nanoparticles for three-dimensional cell culture. Biomaterials. 2013, 34(34):8555-63, doi:10.1016/j.biomaterials.2013.07.056. DOI: https://doi.org/10.1016/j.biomaterials.2013.07.056
- Fürsatz M.; Gerges P.; Wolbank S.; et al. Autonomous spheroid formation by culture plate compartmentation. Biofabrication. 2021, 13(3):035018, doi:10.1088/1758-5090/abe186. DOI: https://doi.org/10.1088/1758-5090/abe186
- Fürsatz M.; Gerges P.; Wolbank S.; et al. A novel system inducing autonomous spheroid formation of cell monolayers. Osteoarthritis and Cartilage. 2021, 29:S407-S8, doi:10.1016/j.joca.2021.02.530. DOI: https://doi.org/10.1016/j.joca.2021.02.530
- Gao M.; Harper M.M.; Lin M.; et al. Development of a Single-Cell Technique to Increase Yield and Use of Gastrointestinal Cancer Organoids for Personalized Medicine Application. J. Am. Coll. Surg., 2021, 232(4):504-14,doi:10.1016/j.jamcollsurg.2020.11.009. DOI: https://doi.org/10.1016/j.jamcollsurg.2020.11.009
- Lee S.Y.; Teng Y.; Son M.; et al. High-dose drug heat map analysis for drug safety and efficacy in multi-spheroid brain normal cells and GBM patient-derived cells. PLoS One. 2021, 16(12):e0251998, doi:10.1371/journal.pone.0251998. DOI: https://doi.org/10.1371/journal.pone.0251998
- Vadivelu R.K.; Ooi C.H.; Yao R.-Q.;et al. Generation of three-dimensional multiple spheroid model of olfactory ensheathing cells using floating liquid marbles. Sci. Rep., 2015, 5(1):15083, doi:10.1038/srep15083. DOI: https://doi.org/10.1038/srep15083
- Chen M.; Shah M.P.; Shelper T.B.; et al. Naked Liquid Marbles: A Robust Three-Dimensional Low-Volume Cell-Culturing System. ACS Appl. Mater. Interfaces. 2019, 11(10):9814-23, doi:10.1021/acsami.8b22036. DOI: https://doi.org/10.1021/acsami.8b22036
- Schindler M.; Siriwardena D.; Kohler T.N.; et al. Agarose microgel culture delineates lumenogenesis in naive and primed human pluripotent stem cells. Stem Cell Reports. 2021, 16(5):1347-62, doi:10.1016/j.stemcr.2021.04.009. DOI: https://doi.org/10.1016/j.stemcr.2021.04.009
- Cui X.; Liu Y.; Hartanto Y.; et al. Multicellular Spheroids Formation and Recovery in Microfluidics-generated Thermoresponsive Microgel Droplets. Coll. Interf. Scie. Comm., 2016, 14:4-7.https://doi.org/10.1016/j.colcom.2016.09.001. DOI: https://doi.org/10.1016/j.colcom.2016.09.001
- Chen D.; Tan Y.; Li Z.; et al. Organoid Cultures Derived From Patients With Papillary Thyroid Cancer. J. Clin. Endocrinol. Metab., 2021, 106(5):1410-26, doi: 10.1210/clinem/dgab020. DOI: https://doi.org/10.1210/clinem/dgab020
- Kim J.; Koo B.-K.; Knoblich J.A. Human organoids: model systems for human biology and medicine. Nat. Rev. Mol. Cell Biol., 2020, 21(10):571-84, doi:10.1038/s41580-020-0259-3. DOI: https://doi.org/10.1038/s41580-020-0259-3
- Sato T.; Vries R.G.; Snippert H.J.; et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009, 459(7244):262-5, doi:10.1038/nature07935. DOI: https://doi.org/10.1038/nature07935
- Takahashi K.; Tanabe K.; Ohnuki M.; et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007, 131(5):861-72, doi:10.1016/j.cell.2007.11.019. DOI: https://doi.org/10.1016/j.cell.2007.11.019
- Spence J.R.; Mayhew C.N.; Rankin S.A.; et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 2011, 470(7332):105-9, doi:10.1038/nature09691. DOI: https://doi.org/10.1038/nature09691
- Wallach T.E.; Bayrer J.R. Intestinal Organoids: New Frontiers in the Study of Intestinal Disease and Physiology. J. Pediatr. Gastroenterol. Nutr., 2017, 64(2):180-5, doi:10.1097/MPG.0000000000001411. DOI: https://doi.org/10.1097/MPG.0000000000001411
- Crespo M.; Vilar E.; Tsai S.Y.; et al. Colonic organoids derived from human induced pluripotent stem cells for modeling colorectal cancer and drug testing. Nat. Med., 2017, 23(7):878-84, doi:10.1038/nm.4355. DOI: https://doi.org/10.1038/nm.4355
- Rodansky E.S.; Johnson L.A.; Huang S.; et al Intestinal organoids: A model of intestinal fibrosis for evaluating anti-fibrotic drugs. Exp. Mol. Pathol., 2015, 98(3):346-51, doi:10.1016/j.yexmp.2015.03.033. DOI: https://doi.org/10.1016/j.yexmp.2015.03.033
- Li V.S.W. Modelling intestinal inflammation and infection using ‘mini-gut’ organoids. Nat. Rev. Gastroenterol. Hepatol., 2021, 18(2):89-90, doi:10.1038/s41575-020-00391-4. DOI: https://doi.org/10.1038/s41575-020-00391-4
- van de Wetering M.; Francies H.E.; Francis J.M.; et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015, 161(4):933-45, doi:10.1016/j.cell.2015.03.053. DOI: https://doi.org/10.1016/j.cell.2015.03.053
- Yin Y.-B.; de Jonge H.R.; Wu X.;et al. Mini-gut: a promising model for drug development. Drug Disc. Today. 2019, 24(9):1784-94, doi:10.1016/j.drudis.2019.06.006. DOI: https://doi.org/10.1016/j.drudis.2019.06.006
- Yoshida S., Miwa H.; Kawachi T.; et al. Generation of intestinal organoids derived from human pluripotent stem cells for drug testing. Sci. Rep., 2020, 10(1):5989, doi:10.1038/s41598-020-63151-z. DOI: https://doi.org/10.1038/s41598-020-63151-z
- Vlachogiannis G.; Hedayat S.; Vatsiou A.; et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science. 2018, 359(6378):920-6, doi:10.1126/science.aao2774. DOI: https://doi.org/10.1126/science.aao2774
- Norkin M.; Ordóñez-Morán P.; Huelsken J. High-content.; targeted RNA-seq screening in organoids for drug discovery in colorectal cancer. Cell Rep., 2021, 35(3):109026, doi:10.1016/j.celrep.2021.109026. DOI: https://doi.org/10.1016/j.celrep.2021.109026
- Raven A.; Lu W.-Y.; Man T.Y.;et al. Cholangiocytes act as facultative liver stem cells during impaired hepatocyte regeneration. Nature. 2017, 547(7663):350-4, doi:10.1038/nature23015. DOI: https://doi.org/10.1038/nature23015
- Huch M.; Dorrell C.; Boj S.F.; et al. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature. 2013, 494(7436):247-50, doi:10.1038/nature11826. DOI: https://doi.org/10.1038/nature11826
- Huch M.; Gehart H.; Van Boxtel R.; et al. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell. 2015, 160(1-2):299-312, doi:10.1016/j.cell.2014.11.050. DOI: https://doi.org/10.1016/j.cell.2014.11.050
- Takebe T.; Sekine K.; Enomura M.; et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature. 2013, 499(7459):481-4, doi:10.1038/nature12271. DOI: https://doi.org/10.1038/nature12271
- Guan Y.; Xu D., Garfin P.M.; et al. Human hepatic organoids for the analysis of human genetic diseases. JCI insight. 2017, 2(17), doi:10.1172/jci.insight.94954. DOI: https://doi.org/10.1172/jci.insight.94954
- Vyas D.; Baptista P.M.; Brovold M.; et al. Self-assembled liver organoids recapitulate hepatobiliary organogenesis in vitro. Hepatology. 2018, 67(2):750-61, doi:10.1002/hep.29483. DOI: https://doi.org/10.1002/hep.29483
- Shinozawa T.; Kimura M.; Cai Y.; et al. High-Fidelity Drug-Induced Liver Injury Screen Using Human Pluripotent Stem Cell–Derived Organoids. Gastroenterology. 2021, 160(3):831-46.e10, doi:10.1053/j.gastro.2020.10.002.
- Mun S.J.; Ryu J .-S.; Lee M-O.;et al. Generation of expandable human pluripotent stem cell-derived hepatocyte-like liver organoids. J. Hepatol., 2019, 71(5):970-85, doi:10.1016/j.jhep.2019.06.030. DOI: https://doi.org/10.1016/j.jhep.2019.06.030
- Shinozawa T.; Kimura M.; Cai Y.; et al. High-fidelity drug-induced liver injury screen using human pluripotent stem cell–derived organoids. Gastroenterology. 2021, 160(3):831-46. e10, doi:10.1053/j.gastro.2020.10.002.
- Schwank G.; Koo B.-K.; Sasselli V.;et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem cell. 2013, 13(6):653-8, doi:10.1016/j.stem.2013.11.002. DOI: https://doi.org/10.1016/j.stem.2013.11.002
- Nie Y.-Z.; Zheng Y.-W.; Miyakawa K.; et al. Recapitulation of hepatitis B virus–host interactions in liver organoids from human induced pluripotent stem cells. EBioMedicine. 2018, 35:114-23, doi:10.1016/j.ebiom.2018.08.014. DOI: https://doi.org/10.1016/j.ebiom.2018.08.014
- Baktash Y.; Madhav A.; Coller K.E.; et al. Single particle imaging of polarized hepatoma organoids upon hepatitis C virus infection reveals an ordered and sequential entry process. Cell Host Microbe. 2018, 23(3):382-94. e5, doi:10.1016/j.chom.2018.02.005. DOI: https://doi.org/10.1016/j.chom.2018.02.005
- De Crignis E.; Hossain T.; Romal S.; et al. Application of human liver organoids as a patient-derived primary model for HBV infection and related hepatocellular carcinoma. eLife. 2021, 10:e60747, doi:10.7554/eLife.60747. DOI: https://doi.org/10.7554/eLife.60747
- Nuciforo S.; Fofana I.; Matter M.S.; et al. Organoid models of human liver cancers derived from tumor needle biopsies. Cell Rep., 2018, 24(5):1363-76, doi:10.1016/j.celrep.2018.07.001 . DOI: https://doi.org/10.1016/j.celrep.2018.07.001
- Li L.; Knutsdottir H.; Hui K.; et al. Human primary liver cancer organoids reveal intratumor and interpatient drug response heterogeneity. JCI insight. 2019, 4(2), doi:10.1172/jci.insight.121490. DOI: https://doi.org/10.1172/jci.insight.121490
- Broutier L.; Mastrogiovanni G.; Verstegen M.; et al. Human primary liver cancer–derived organoid cultures for disease modeling and drug screening. Nat. Med., 2017, 23(12):1424-35, doi:10.1038/nm.4438. DOI: https://doi.org/10.1038/nm.4438
- Artegiani B.; van Voorthuijsen L.; Lindeboom R.G.; et al. Probing the tumor suppressor function of BAP1 in CRISPR-engineered human liver organoids. Cell Stem Cell. 2019, 24(6):927-43. e6, doi:10.1016/j.stem.2019.04.017. DOI: https://doi.org/10.1016/j.stem.2019.04.017
- Khetani S.R. Pluripotent Stem Cell-Derived Human Liver Organoids Enter the Realm of High-Throughput Drug Screening. Gastroenterology. 2021, 160(3):653-5, doi:10.1053/j.gastro.2020.12.005 . DOI: https://doi.org/10.1053/j.gastro.2020.12.005
- Shinozawa T.; Kimura M.; Cai Y.; et al. High-Fidelity Drug-Induced Liver Injury Screen Using Human Pluripotent Stem Cell-Derived Organoids. Gastroenterology. 2021, 160(3):831-46 e10, doi:10.1053/j.gastro.2020.10.002. DOI: https://doi.org/10.1053/j.gastro.2020.10.002
- Huang L.; Holtzinger A.; Jagan I.; et al. Ductal pancreatic cancer modeling and drug screening using human pluripotent stem cell- and patient-derived tumor organoids. Nat Med. 2015, 21(11):1364-71, doi:10.1038/nm.3973. DOI: https://doi.org/10.1038/nm.3973
- Greggio C.; De Franceschi F.; Figueiredo-Larsen M.; et al. Artificial three-dimensional niches deconstruct pancreas development in vitro. Development. 2013, 140(21):4452-62, doi:10.1242/dev.096628. DOI: https://doi.org/10.1242/dev.096628
- Grapin-Botton A. Three-dimensional pancreas organogenesis models. Diabetes Obes. Metab., 2016, 18(S1):33-40, doi:10.1111/dom.12720. DOI: https://doi.org/10.1111/dom.12720
- Nostro M.C.; Sarangi F.; Yang C.; et al. Efficient generation of NKX6-1+ pancreatic progenitors from multiple human pluripotent stem cell lines. Stem Cell Reports. 2015, 4(4):591-604, doi:10.1016/j.stemcr.2015.02.017. DOI: https://doi.org/10.1016/j.stemcr.2015.02.017
- Hohwieler M.; Illing A.; Hermann P.C.; et al. Human pluripotent stem cell-derived acinar/ductal organoids generate human pancreas upon orthotopic transplantation and allow disease modelling. Gut. 2017, 66(3):473-86, doi:10.1136/gutjnl-2016-312423. DOI: https://doi.org/10.1136/gutjnl-2016-312423
- Dahl-Jensen S.B.; Figueiredo-Larsen M.; Grapin-Botton A.; et al. Short-range growth inhibitory signals from the epithelium can drive non-stereotypic branching in the pancreas. Phys. Biol., 2016, 13(1):016007, doi:10.1088/1478-3975/13/1/016007. DOI: https://doi.org/10.1088/1478-3975/13/1/016007
- Driehuis E.; Gracanin A.; Vries R.G.J.; Clevers H , Boj S.F. Establishment of Pancreatic Organoids from Normal Tissue and Tumors. STAR Protoc., 2020, 1(3):100192, doi:10.1016/j.xpro.2020.100192. DOI: https://doi.org/10.1016/j.xpro.2020.100192
- Tiriac H.; Belleau P.; Engle D.D.; et al. Organoid Profiling Identifies Common Responders to Chemotherapy in Pancreatic Cancer. Cancer Discov., 2018, 8(9):1112-29, doi:10.1158/2159-8290.CD-18-0349. DOI: https://doi.org/10.1158/2159-8290.CD-18-0349
- Hirt C.K.; Booij T.H.; Grob L.; et al. Drug screening and genome editing in human pancreatic cancer organoids identifies drug-gene interactions and candidates for off-label therapy. Cell Genomics. 2022, 2(2):100095, doi:10.1016/j.xgen.2022.100095. DOI: https://doi.org/10.1016/j.xgen.2022.100095
- Hou S.; Tiriac H.; Sridharan B.P.; et al. Advanced Development of Primary Pancreatic Organoid Tumor Models for High-Throughput Phenotypic Drug Screening. SLAS DISC:, 2018, 23(6):574-84.doi:10.1177/2472555218766842. DOI: https://doi.org/10.1177/2472555218766842
- Shi X.; Li Y.; Yuan Q.; et al. Integrated profiling of human pancreatic cancer organoids reveals chromatin accessibility features associated with drug sensitivity. Nat. Commu., 2022, 13(1):2169.doi:10.1038/s41467-022-29857-6. DOI: https://doi.org/10.1038/s41467-022-29857-6
- Yin J.; Meng H.; Lin J.; et al. Pancreatic islet organoids-on-a-chip: how far have we gone? J. Nanobiotechnology. 2022, 20(1):308, doi:10.1186/s12951-022-01518-2. DOI: https://doi.org/10.1186/s12951-022-01518-2
- Lancaster M.A.; Renner M.; Martin C.-A.;et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013, 501(7467):373-9, doi:10.1038/nature12517. DOI: https://doi.org/10.1038/nature12517
- Cho A.-N.; Jin Y.; An Y.;et al. Microfluidic device with brain extracellular matrix promotes structural and functional maturation of human brain organoids. Nat. Commun., 2021, 12(1):4730, doi:10.1038/s41467-021-24775-5. DOI: https://doi.org/10.1038/s41467-021-24775-5
- Lee C.-T.; Bendriem R.M.; Wu W.W.; et al. 3D brain Organoids derived from pluripotent stem cells: promising experimental models for brain development and neurodegenerative disorders. Journal of Biomedical Science. 2017, 24(1):59, doi:10.1186/s12929-017-0362-8. DOI: https://doi.org/10.1186/s12929-017-0362-8
- Ao Z.; Cai H.; Wu Z.; et al . Controllable fusion of human brain organoids using acoustofluidics. Lab Chip. 2021, 21(4):688-99, doi:10.1039/d0lc01141j. DOI: https://doi.org/10.1039/D0LC01141J
- Di Lullo E.; Kriegstein A.R. The use of brain organoids to investigate neural development and disease. Nat. Rev. Neurosci., 2017, 18(10):573-84, doi:10.1038/nrn.2017.107. DOI: https://doi.org/10.1038/nrn.2017.107
- Luo J. ; Li P . Human pluripotent stem cell-derived brain organoids as in vitro models for studying neural disorders and cancer. Cell Bios., 2021, 11(1):99, doi:10.1186/s13578-021-00617-1. DOI: https://doi.org/10.1186/s13578-021-00617-1
- Qian X.; Nguyen H.N.; Jacob F.; et al. Using brain organoids to understand Zika virus-induced microcephaly. Development. 2017, 144(6):952-7, doi:10.1242/dev.140707. DOI: https://doi.org/10.1242/dev.140707
- Gabriel E.; Ramani A.; Karow U.; et al. Recent Zika Virus Isolates Induce Premature Differentiation of Neural Progenitors in Human Brain Organoids. Cell Stem Cell. 2017, 20(3):397-406.e5, doi:10.1016/j.stem.2016.12.005. DOI: https://doi.org/10.1016/j.stem.2016.12.005
- Lee S.-E.; Shin N.; Kook M.G.;et al. Human iNSC-derived brain organoid model of lysosomal storage disorder in Niemann–Pick disease type C. Cell Death Dis., 2020, 11(12):1059.doi:10.1038/s41419-020-03262-7. DOI: https://doi.org/10.1038/s41419-020-03262-7
- Zhang L.; Liu F.; Weygant N.; et al. A novel integrated system using patient-derived glioma cerebral organoids and xenografts for disease modeling and drug screening. Cancer Lett., 2021, 500:87-97.doi:10.1016/j.canlet.2020.12.013. DOI: https://doi.org/10.1016/j.canlet.2020.12.013
- Goranci-Buzhala G.; Mariappan A.; Gabriel E.; et al. Rapid and Efficient Invasion Assay of Glioblastoma in Human Brain Organoids. Cell Rep., 2020, 31(10):107738, doi:10.1016/j.celrep.2020.107738. DOI: https://doi.org/10.1016/j.celrep.2020.107738
- Mansour A.A.; Gonçalves J.T.; Bloyd C.W.; et al. An in vivo model of functional and vascularized human brain organoids. Nature Biotechnology. 2018, 36(5):432-41, doi:10.1038/nbt.4127. DOI: https://doi.org/10.1038/nbt.4127
- Tran H .N; Gautam V. Micro- and nanodevices for integration with human brain organoids. Biosens. Bioelectro., 2022, 114734. doi:1016/j.bios.2022.114734 DOI: https://doi.org/10.1016/j.bios.2022.114734
- Fischer J.; Heide M.; Huttner W.B. Genetic Modification of Brain Organoids. Front. Cell. Neurosci., 2019, 13, doi:10.3389/fncel.2019.00558. DOI: https://doi.org/10.3389/fncel.2019.00558
- Durens M.; Nestor J.; Williams M.; et al. High-throughput screening of human induced pluripotent stem cell-derived brain organoids. Journal of Neuroscience Methods. 2020, 335:108627. DOI: https://doi.org/10.1016/j.jneumeth.2020.108627
- Park J.-C.; Jang S.-Y.; Lee D.; et al. A logical network-based drug-screening platform for Alzheimer’s disease representing pathological features of human brain organoids. Nat. Commu., 2021, 12(1):280.doi:10.1038/s41467-020-20440-5. DOI: https://doi.org/10.1038/s41467-020-20440-5
- Nestor M.W.; Paull D.; Jacob S.; et al. Differentiation of serum-free embryoid bodies from human induced pluripotent stem cells into networks. Stem Cell Res. 2013, 10(3):454-63.doi:10.1016/j.scr.2013.02.001. DOI: https://doi.org/10.1016/j.scr.2013.02.001
- Brennand K.J.; Marchetto M.C.; Benvenisty N.; et al. Creating Patient-Specific Neural Cells for the In Vitro Study of Brain Disorders. Stem Cell Reports. 2015, 5(6):933-45, doi:10.1016/j.stemcr.2015.10.011. DOI: https://doi.org/10.1016/j.stemcr.2015.10.011
- Nugraha B.; Buono M.F.; von Boehmer L.; et al. Human Cardiac Organoids for Disease Modeling. Clin. Pharmacol. Ther., 2019, 105(1):79-85.doi:10.1002/cpt.1286. DOI: https://doi.org/10.1002/cpt.1286
- Beauchamp P.; Jackson C.B.; Ozhathil L.C.; et al. 3D Co-culture of hiPSC-Derived Cardiomyocytes With Cardiac Fibroblasts Improves Tissue-Like Features of Cardiac Spheroids. Front. Mol. Biosci., 2020, 7.doi:10.3389/fmolb.2020.00014. DOI: https://doi.org/10.3389/fmolb.2020.00014
- Filippo Buono M.; von Boehmer L.; Strang J.; et al. Human Cardiac Organoids for Modeling Genetic Cardiomyopathy. Cells. 2020, 9(7):1733, doi:10.3390/cells9071733. DOI: https://doi.org/10.3390/cells9071733
- Richards D.J.; Li Y.; Kerr C.M.; et al. Human cardiac organoids for the modelling of myocardial infarction and drug cardiotoxicity. Nat. Biomed. Eng., 2020, 4(4):446-62, doi:10.1038/s41551-020-0539-4. DOI: https://doi.org/10.1038/s41551-020-0539-4
- Skardal A.; Shupe T.; Atala A. Organoid-on-a-chip and body-on-a-chip systems for drug screening and disease modeling. Drug Discov. Today. 2016, 21(9):1399-411.doi:10.1016/j.drudis.2016.07.003. DOI: https://doi.org/10.1016/j.drudis.2016.07.003
- Forsythe S.D.; Devarasetty M.; Shupe T.; et al. Environmental Toxin Screening Using Human-Derived 3D Bioengineered Liver and Cardiac Organoids. Front. Public. Health. 2018, 6, doi:10.3389/fpubh.2018.00103. DOI: https://doi.org/10.3389/fpubh.2018.00103
- Hoang P.; Kowalczewski A.; Sun S.; et al. Engineering spatial-organized cardiac organoids for developmental toxicity testing. Stem Cell Reports. 2021, 16(5):1228-44, doi:10.1016/j.stemcr.2021.03.013. DOI: https://doi.org/10.1016/j.stemcr.2021.03.013
- Mills R.J.; Titmarsh D.M.; Koenig X.; et al. Functional screening in human cardiac organoids reveals a metabolic mechanism for cardiomyocyte cell cycle arrest. Proc Natl Acad Sci U S A. 2017, 114(40):E8372-E81, doi:10.1073/pnas.1707316114. DOI: https://doi.org/10.1073/pnas.1707316114
- Zhang Y.S.; Arneri A.; Bersini S.; et al. Bioprinting 3D microfibrous scaffolds for engineering endothelialized myocardium and heart-on-a-chip. Biomaterials. 2016, 110:45-59.doi:10.1016/j.biomaterials.2016.09.003. DOI: https://doi.org/10.1016/j.biomaterials.2016.09.003
- Cho J.; Lee H.; Rah W.; et al. From engineered heart tissue to cardiac organoid. Theranostics. 2022, 12(6):2758-72.doi:10.7150/thno.67661. DOI: https://doi.org/10.7150/thno.67661
- Wimmer R.A.; Leopoldi A.; Aichinger M.; et al. Human blood vessel organoids as a model of diabetic vasculopathy. Nature. 2019, 565(7740):505-10, doi:10.1038/s41586-018-0858-8. DOI: https://doi.org/10.1038/s41586-018-0858-8
- Kim H.; Kamm R.D.; Vunjak-Novakovic G.; et al. Progress in multicellular human cardiac organoids for clinical applications. Cell Stem Cell. 2022, 29(4):503-14, doi:10.1016/j.stem.2022.03.012. DOI: https://doi.org/10.1016/j.stem.2022.03.012
- Drakhlis L.; Biswanath S.; Farr C.-M.;et al. Human heart-forming organoids recapitulate early heart and foregut development. Nat. Biotechnol., 2021, 39(6):737-46, doi:10.1038/s41587-021-00815-9. DOI: https://doi.org/10.1038/s41587-021-00815-9
- Drakhlis L.; Devadas S.B.; Zweigerdt R. Generation of heart-forming organoids from human pluripotent stem cells. Nat. Protoc., 2021, 16(12):5652-72, doi:10.1038/s41596-021-00629-8. DOI: https://doi.org/10.1038/s41596-021-00629-8
- Lewis-Israeli Y.R.; Wasserman A.H.; Gabalski M.A.; et al. Self-assembling human heart organoids for the modeling of cardiac development and congenital heart disease. Nat Commun. 2021, 12(1):5142, doi:10.1038/s41467-021-25329-5. DOI: https://doi.org/10.1038/s41467-021-25329-5
- Hofbauer P.; Jahnel S.M.; Papai N.; et al. Cardioids reveal self-organizing principles of human cardiogenesis. Cell. 2021, 184(12):3299-317 e22, doi:10.1016/j.cell.2021.04.034 . DOI: https://doi.org/10.1016/j.cell.2021.04.034
- Lee J.; Sutani A.; Kaneko R. ; et al. In vitro generation of functional murine heart organoids via FGF4 and extracellular matrix. Nat. Commu., 2020, 11(1):4283.doi:10.1038/s41467-020-18031-5. DOI: https://doi.org/10.1038/s41467-020-18031-5
- Rossi G.; Broguiere N.; Miyamoto M.; et al. Capturing Cardiogenesis in Gastruloids. Cell Stem Cell. 2021, 28(2):230-40 e6, doi:10.1016/j.stem.2020.10.013 . DOI: https://doi.org/10.1016/j.stem.2020.10.013
- Lewis-Israeli Y.R.; Abdelhamid M. ; Olomu I.; et al. Modeling the Effects of Maternal Diabetes on the Developing Human Heart Using Pluripotent Stem Cell–Derived Heart Organoids. Curr. Protoc., 2022, 2(6):e461, doi:10.1002/cpz1.461. DOI: https://doi.org/10.1002/cpz1.461
- Skardal A.; Aleman J.; Forsythe S.; et al. Drug compound screening in single and integrated multi-organoid body-on-a-chip systems. Biofabrication. 2020, 12(2):025017, doi:10.1088/1758-5090/ab6d36. DOI: https://doi.org/10.1088/1758-5090/ab6d36
- Park S.E.; Georgescu A.; Huh D. Organoids-on-a-chip. Science. 2019, 364(6444):960-5, doi:10.1126/science.aaw7894. DOI: https://doi.org/10.1126/science.aaw7894
- Ferrari E.; Rasponi M. Liver–Heart on chip models for drug safety. APL Bioengineering. 2021, 5(3):031505, doi:10.1063/5.0048986. DOI: https://doi.org/10.1063/5.0048986
- Parlato S.; Grisanti G.; Sinibaldi G.; et al. Tumor-on-a-chip platforms to study cancer–immune system crosstalk in the era of immunotherapy. Lab Chip. 2021, 21(2):234-53, doi:10.1039/d0lc00799d. DOI: https://doi.org/10.1039/D0LC00799D





