Autophagy is a conserved degradative process which facilitates the degradation of excessive or damaged proteins and organelles in lysosomes. It plays a crucial role in maintaining cellular homeostasis by liberating glucose and other nutrients, like amino acids and fatty acids, to support cellular functions during stress conditions and starvation. Glucose, an important carbohydrate in the human body sustains cellular life. Recent studies have highlighted the role of autophagy in glucose metabolism. In this review, we summarize the role of autophagy in glycogen metabolism, glycolysis, and gluconeogenesis. We also explore the relationship between autophagy and factors involved in glucose metabolism, such as ATP and calcium. Additionally, we discuss the effects of autophagy on diseases associated with abnormal glucose metabolism, including diabetes and insulin resistance (IR). Furthermore, we will provide an overview of potential medications that can improve glucose metabolism by regulating autophagy. Thus, as the main regulator, autophagy is promising as a therapeutic target for addressing abnormal glucose metabolism-related diseases.
- Open Access
- Review
Autophagy: A Major Regulatory Factor in Glucose Metabolism
- Meiqing Liu 1, 2, †,
- Xuling Luo 2, †,
- Haoliang Hu 3, *,
- Linxi Chen 2, *
Author Information
Received: 23 Dec 2024 | Revised: 03 Jan 2025 | Accepted: 02 Apr 2025 | Published: 04 Aug 2025
Abstract
Graphical Abstract
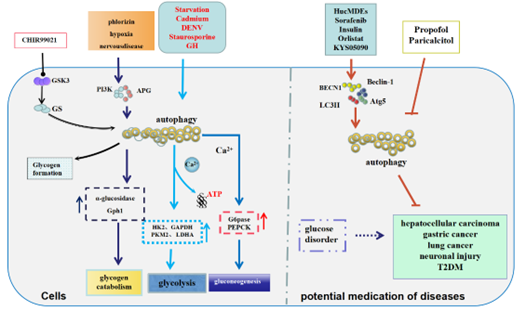
Keywords
autophagy | glycogen | glycolysis | gluconeogenesis | diabetes | potential medication
References
- 1.Poggiogalle, E.; Jamshed, H.; Peterson, C.M. Circadian regulation of glucose, lipid, and energy metabolism in humans. Metabolism 2018, 84, 11–27. https://doi.org/10.1016/j.metabol.2017.11.017.
- 2.Sajadimajd, S.; Bahrami, G.; Mohammadi, B.; Nouri, Z.; Farzaei, M.H.; Chen, J.T. Protective effect of the isolated oligosaccharide from Rosa canina in STZ-treated cells through modulation of the autophagy pathway. J. Food Biochem. 2020, 44, e13404. https://doi.org/10.1111/jfbc.13404.
- 3.Han, H.S.; Kang, G.; Kim, J.S.; Choi, B.H.; Koo, S.H. Regulation of glucose metabolism from a liver-centric perspective. Exp. Mol. Med. 2016, 48, e218. https://doi.org/10.1038/emm.2015.122.
- 4.Hernandez, F. Glycolysis and gluconeogenesis: A teaching view. J. Biol. Chem. 2021, 296, 100016. https://doi.org/10.1016/j.jbc.2020.100016.
- 5.Huang, F.; Wang, B.R.; Wang, Y.G. Role of autophagy in tumorigenesis, metastasis, targeted therapy and drug resistance of hepatocellular carcinoma. World J. Gastroenterol. 2018, 24, 4643–4651. https://doi.org/10.3748/wjg.v24.i41.4643.
- 6.Levine, B.; Klionsky, D.J. Development by self-digestion: Molecular mechanisms and biological functions of autophagy. Dev. Cell 2004, 6, 463–477. https://doi.org/10.1016/s1534-5807(04)00099-1.
- 7.McCarthy, M. Japanese cellular biologist wins Nobel prize for study of autophagy. BMJ 2016, 355, i5374. https://doi.org/10.1136/bmj.i5374.
- 8.Levine, B.; Klionsky, D. Autophagy wins the 2016 Nobel Prize in Physiology or Medicine: Breakthroughs in baker’s yeast fuel advances in biomedical research. Proc. Natl. Acad. Sci. USA 2017, 114, 201–205. https://doi.org/10.1073/pnas.1619876114.
- 9.Mizushima, N.; Komatsu, M. Autophagy: Renovation of cells and tissues. Cell 2011, 147, 728–741. https://doi.org/10.1016/j.cell.2011.10.026.
- 10.Hurley, J.H.; Young, L.N. Mechanisms of Autophagy Initiation. Annu. Rev. Biochem. 2017, 86, 225–244. https://doi.org/10.1146/annurev-biochem-061516-044820.
- 11.Chu, C.T. Mechanisms of selective autophagy and mitophagy: Implications for neurodegenerative diseases. Neurobiol. Dis. 2019, 122, 23–34. https://doi.org/10.1016/j.nbd.2018.07.015.
- 12.Luo, Z.; Xu, X.; Sho, T.; Zhang, J.; Xu, W.; Yao, J.; Xu, J. ROS-induced autophagy regulates porcine trophectoderm cell apoptosis, proliferation, and differentiation. Am. J. Physiol. Cell Physiol. 2019, 316, C198–c209. https://doi.org/10.1152/ajpcell.00256.2018.
- 13.Qi, Z.; Chen, L. Endoplasmic Reticulum Stress and Autophagy. Adv. Exp. Med. Biol. 2019, 362, 11. https://doi.org/10.1007/978-981-15-0602-4_8.
- 14.Liu, M.; Li, H.; Zhou, Q.; Zhao, H.; Lv, D.; Cao, J.; Jiang, J.; Tang, M.; Wu, D.; Liu, J.; et al. ROS-Autophagy pathway mediates monocytes-human umbilical vein endothelial cells adhesion induced by apelin-13. J. Cell. Physiol. 2018, 233, 6839–6850. https://doi.org/10.1002/jcp.26554.
- 15.Xie, F.; Liu, W.; Feng, F.; Li, X.; He, L.; Lv, D.; Qin, X.; Li, L.; Li, L.; Chen, L. Apelin-13 promotes cardiomyocyte hypertrophy via PI3K-Akt-ERK1/2-p70S6K and PI3K-induced autophagy. Acta Biochim. Biophys. Sin. 2015, 47, 969–980. https://doi.org/10.1093/abbs/gmv111.
- 16.Zhe, C.; Jun, C.; Qun, Z.; Le-le, W.; Jia-Wei, C.; Xiang-Ning, D.; Jia-Long, Y.; Jian-Gang, C.; Xiao-Dan, X.; Lan-Fang, L.; et al. SEC62-dependent ER-phagy contributes to apelin-13/APJ-induced monocyte-vascular endothelial cell adhesion in atherosclerosis pathogenesis. Acta Pharmacol. Sin. 2025, 46, 1652–1663. https://doi.org/10.1038/s41401-024-01471-w.
- 17.Boya, P.; Codogno, P.; Rodriguez-Muela, N. Autophagy in stem cells: Repair, remodelling and metabolic reprogramming. Development 2018, 145, dev146506. https://doi.org/10.1242/dev.146506.
- 18.Butler, D.E.; Marlein, C.; Walker, H.F.; Frame, F.M.; Mann, V.M.; Simms, M.S.; Davies, B.R.; Collins, A.T.; Maitland, N.J. Inhibition of the PI3K/AKT/mTOR pathway activates autophagy and compensatory Ras/Raf/MEK/ERK signalling in prostate cancer. Oncotarget 2017, 8, 56698–56713. https://doi.org/10.18632/oncotarget.18082.
- 19.Qian, H.; Chao, X.; Williams, J.; Fulte, S.; Li, T.; Yang, L.; Ding, W.X. Autophagy in liver diseases: A review. Mol. Asp. Med. 2021, 82, 100973. https://doi.org/10.1016/j.mam.2021.100973.
- 20.Mikhail, R.; Anna, E.; Deborah, S.; Wenfang, X.; Eitan, O.; Samuel, B.; Petra, E.G.; Reid, R.T.; William, S.L.; Tamas, D.; et al. Diurnal Rhythms Spatially and Temporally Organize Autophagy. Cell Rep. 2019, 26, 1880-1892. https://doi.org/10.1016/j.celrep.2019.01.072.
- 21.Martinez-Lopez, N.; Singh, R. Autophagy and Lipid Droplets in the Liver. Annu. Rev. Nutr. 2015, 35, 215–237. https://doi.org/10.1146/annurev-nutr-071813-105336.
- 22.Filali-Mouncef, Y.; Hunter, C.; Roccio, F.; Zagkou, S.; Dupont, N.; Primard, C.; Proikas-Cezanne, T.; Reggiori, F. The ménage à trois of autophagy, lipid droplets and liver disease. Autophagy 2022, 18, 50–72. https://doi.org/10.1080/15548627.2021.1895658.
- 23.Kalamidas, S.; Kotoulas, O.; Kotoulas, A.; Maintas, D. The breakdown of glycogen in the lysosomes of newborn rat hepatocytes: The effects of glucose, cyclic 3′,5′-AMP and caffeine. Histol. Histopathol. 1994, 9, 691–698.
- 24.Kotoulas, O.B.; Ho, J.; Adachi, F.; Weigensberg, B.I.; Phillips, M.J. Fine structural aspects of the mobilization of hepatic glycogen. II. Inhibition of glycogen breakdown. Am. J. Pathol. 1971, 63, 23–36.
- 25.Kotoulas, O.B.; Phillips, M.J. Fine structural aspects of the mobilization of hepatic glycogen. I. Acceleration of glycogen breakdown. Am. J. Pathol. 1971, 63, 1–22.
- 26.Roach, P.J.; Depaoli-Roach, A.A.; Hurley, T.D.; Tagliabracci, V.S. Glycogen and its metabolism: Some new developments and old themes. Biochem. J. 2012, 441, 763–787. https://doi.org/10.1042/bj20111416.
- 27.Kalamidas, S.A.; Kotoulas, O.B. Studies on the breakdown of glycogen in the lysosomes: The effects of hydrocortisone. Histol. Histopathol. 2000, 15, 29–35. https://doi.org/10.14670/hh-15.29.
- 28.Kalamidas, S.A.; Kotoulas, O.B. The degradation of glycogen in the lysosomes of newborn rat hepatocytes: Glycogen-, maltose- and isomaltose-hydrolyzing acid alpha glucosidase activities in liver. Histol. Histopathol. 1999, 14, 23–30. https://doi.org/10.14670/hh-14.23.
- 29.Iwamasa, T.; Tsuru, T.; Hamada, T.; Takeuchi, T. Physicochemical and ultrastructural studies on glycogenosomes in newborn rat hepatocytes. Pathol. Res. Pract. 1980, 167, 363–373. https://doi.org/10.1016/s0344-0338(80)80065-3.
- 30.Zhao, H.; Tang, M.; Liu, M.; Chen, L. Glycophagy: An emerging target in pathology. Clin. Chim. Acta 2018, 484, 298–303. https://doi.org/10.1016/j.cca.2018.06.014.
- 31.Dong, J.; Guo, C.; Yang, Z.; Wu, Y.; Zhang, C. Follicle-Stimulating Hormone Alleviates Ovarian Aging by Modulating Mitophagy- and Glycophagy-Based Energy Metabolism in Hens. Cells 2022, 11, 3270. https://doi.org/10.3390/cells11203270.
- 32.Qiu, F.; Yuan, Y.; Luo, W.; Gong, Y.; Zhang, Z.; Liu, Z.M.; Gao, L. Asiatic acid alleviates ischemic myocardial injury in mice by modulating mitophagy- and glycophagy-based energy metabolism. Acta Pharmacol. Sin. 2021, 43, 1395–1407. https://doi.org/10.1038/s41401-021-00763-9.
- 33.Kalamidas, S.A.; Kotoulas, O.B. Glycogen autophagy in newborn rat hepatocytes. Histol. Histopathol. 2000, 15, 1011–1018. https://doi.org/10.14670/hh-15.1011.
- 34.David, H.; Reinke, P.; Bimmler, M. Postnatal development of hepatocytes following oxygen deficiency in utero. Exp. Pathol. 1986, 30, 247–256. https://doi.org/10.1016/s0232-1513(86)80084-6.
- 35.Devos, P.; Hers, H. Random, presumably hydrolytic, and lysosomal glycogenolysis in the livers of rats treated with phlorizin and of newborn rats. Biochem. J. 1980, 192, 177–181. https://doi.org/10.1042/bj1920177.
- 36.Kovács, A.L.; Eldib, A.; Telbisz, A. Autophagy in hepatocytes and erythropoietic cells isolated from the twenty-one day old rat embryo. Acta Biol. Hung. 2001, 52, 417–433. https://doi.org/10.1556/ABiol.52.2001.4.7.
- 37.Ohshita, T. Suppression of autophagy by ethionine administration in male rat liver in vivo. Toxicology 2000, 147, 51–57. https://doi.org/10.1016/s0300-483x(00)00180-3.
- 38.Lin, Q.; Shi, Y.; Liu, Z.; Mehrpour, M.; Hamaï, A.; Gong, C. Non-coding RNAs as new autophagy regulators in cancer progression. Biochim. Biophys. Acta. Mol. Basis Dis. 2022, 1868, 166293. https://doi.org/10.1016/j.bbadis.2021.166293.
- 39.Deng, Y.Z.; Ramos-Pamplona, M.; Naqvi, N.I. Autophagy-assisted glycogen catabolism regulates asexual differentiation in Magnaporthe oryzae. Autophagy 2009, 5, 33–43. https://doi.org/10.4161/auto.5.1.7175.
- 40.Yang, C.; Wang, H.; Shao, M.; Chu, F.; He, Y.; Chen, X.; Fan, J.; Chen, J.; Cai, Q.; Wu, C. Brain-Type Glycogen Phosphorylase (PYGB) in the Pathologies of Diseases: A Systematic Review. Cells 2024, 13, 289. https://doi.org/10.3390/cells13030289.
- 41.Franco-Romero, A.; Sandri, M.; Schiaffino, S. Autophagy in Skeletal Muscle. Cold Spring Harb. Perspect. Biol. 2024, a041565. https://doi.org/10.1101/cshperspect.a041565.
- 42.Müller, M.S.; Pedersen, S.E.; Walls, A.B.; Waagepetersen, H.S.; Bak, L.K. Isoform-selective regulation of glycogen phosphorylase by energy deprivation and phosphorylation in astrocytes. Glia 2015, 63, 154–162. https://doi.org/10.1002/glia.22741.
- 43.Zois, C.E.; Hendriks, A.M.; Haider, S.; Pires, E.; Bridges, E.; Kalamida, D.; Voukantsis, D.; Lagerholm, B.C.; Fehrmann, R.S.N.; den Dunnen, W.F.A.; et al. Liver glycogen phosphorylase is upregulated in glioblastoma and provides a metabolic vulnerability to high dose radiation. Cell Death Dis. 2022, 13, 573. https://doi.org/10.1038/s41419-022-05005-2.
- 44.Chen, H.; Zhao, F.; Chen, K.; Guo, Y.; Liang, Y.; Zhao, H.; Chen, S. Exposure of zebrafish to a cold environment triggered cellular autophagy in zebrafish liver. J. Fish. Dis. 2022, 45, 991–1000. https://doi.org/10.1111/jfd.13620.
- 45.Schulz, A.; Sekine, Y.; Oyeyemi, M.J.; Abrams, A.J.; Basavaraju, M.; Han, S.M.; Groth, M.; Morrison, H.; Strittmatter, S.M.; Hammarlund, M. The stress-responsive gene GDPGP1/mcp-1 regulates neuronal glycogen metabolism and survival. J. Cell Biol. 2020, 219, e201807127. https://doi.org/10.1083/jcb.201807127.
- 46.Onkar, A.; Sheshadri, D.; Ganesh, S. Glycogen: The missing link in neuronal autophagy? Autophagy 2020, 16, 2102–2104. https://doi.org/10.1080/15548627.2020.1802090.
- 47.Adler, L.; Gomez, T.; Clarke, S.; Linster, C. A novel GDP-D-glucose phosphorylase involved in quality control of the nucleoside diphosphate sugar pool in Caenorhabditis elegans and mammals. J. Biol. Chem. 2011, 286, 21511–21523. https://doi.org/10.1074/jbc.M111.238774.
- 48.Li, I.H.; Ma, K.H.; Weng, S.J.; Huang, S.S.; Liang, C.M.; Huang, Y.S. Autophagy activation is involved in 3,4-methylenedioxymethamphetamine (‘ecstasy’)—induced neurotoxicity in cultured cortical neurons. PLoS ONE 2014, 9, e116565. https://doi.org/10.1371/journal.pone.0116565.
- 49.Zirin, J.; Nieuwenhuis, J.; Perrimon, N. Role of autophagy in glycogen breakdown and its relevance to chloroquine myopathy. PLoS Biol. 2013, 11, e1001708. https://doi.org/10.1371/journal.pbio.1001708.
- 50.Embi, N.; Rylatt, D.B.; Cohen, P. Glycogen synthase kinase-3 from rabbit skeletal muscle. Separation from cyclic-AMP-dependent protein kinase and phosphorylase kinase. Eur. J. Biochem. 1980, 107, 519–527.
- 51.Pan, H.Y.; Valapala, M. Regulation of Autophagy by the Glycogen Synthase Kinase-3 (GSK-3) Signaling Pathway. Int. J. Mol. Sci. 2022, 23, 1709. https://doi.org/10.3390/ijms23031709.
- 52.Cline, G.W.; Johnson, K.; Regittnig, W.; Perret, P.; Tozzo, E.; Xiao, L.; Damico, C.; Shulman, G.I. Effects of a novel glycogen synthase kinase-3 inhibitor on insulin-stimulated glucose metabolism in Zucker diabetic fatty (fa/fa) rats. Diabetes 2002, 51, 2903–2910. https://doi.org/10.2337/diabetes.51.10.2903.
- 53.Patel, S.; Doble, B.W.; MacAulay, K.; Sinclair, E.M.; Drucker, D.J.; Woodgett, J.R. Tissue-specific role of glycogen synthase kinase 3beta in glucose homeostasis and insulin action. Mol. Cell Biol. 2008, 28, 6314–6328. https://doi.org/10.1128/mcb.00763-08.
- 54.Yang, J.; Takahashi, Y.; Cheng, E.; Liu, J.; Terranova, P.F.; Zhao, B.; Thrasher, J.B.; Wang, H.G.; Li, B. GSK-3beta promotes cell survival by modulating Bif-1-dependent autophagy and cell death. J. Cell Sci. 2010, 123, 861–870. https://doi.org/10.1242/jcs.060475.
- 55.Wang, H.; Brown, J.; Gu, Z.; Garcia, C.A.; Liang, R.; Alard, P.; Beurel, E.; Jope, R.S.; Greenway, T.; Martin, M. Convergence of the mammalian target of rapamycin complex 1- and glycogen synthase kinase 3-β-signaling pathways regulates the innate inflammatory response. J. Immunol. 2011, 186, 5217–5226. https://doi.org/10.4049/jimmunol.1002513.
- 56.Weikel, K.A.; Cacicedo, J.M.; Ruderman, N.B.; Ido, Y. Knockdown of GSK3β increases basal autophagy and AMPK signalling in nutrient-laden human aortic endothelial cells. Biosci. Rep. 2016, 36, e00382. https://doi.org/10.1042/bsr20160174.
- 57.Russi, S.; Sgambato, A.; Bochicchio, A.M.; Zoppoli, P.; Aieta, M.; Capobianco, A.M.L.; Ruggieri, V.; Zifarone, E.; Falco, G.; Laurino, S. CHIR99021, trough GSK-3β Targeting, Reduces Epithelioid Sarcoma Cell Proliferation by Activating Mitotic Catastrophe and Autophagy. Int. J. Mol. Sci. 2021, 22, 11147. https://doi.org/10.3390/ijms222011147.
- 58.Ryu, H.Y.; Kim, L.E.; Jeong, H.; Yeo, B.K.; Lee, J.W.; Nam, H.; Ha, S.; An, H.K.; Park, H.; Jung, S.; et al. GSK3B induces autophagy by phosphorylating ULK1. Exp. Mol. Med. 2021, 53, 369–383. https://doi.org/10.1038/s12276-021-00570-6.
- 59.Akram, M. Mini-review on glycolysis and cancer. J. Cancer Educ. 2013, 28, 454–457. https://doi.org/10.1007/s13187-013-0486-9.
- 60.Keenan, M.M.; Chi, J.T. Alternative fuels for cancer cells. Cancer J. 2015, 21, 49–55. https://doi.org/10.1097/PPO.000000000
- 61.Lock, R.; Roy, S.; Kenific, C.M.; Su, J.S.; Salas, E.; Ronen, S.M.; Debnath, J. Autophagy facilitates glycolysis during Ras-mediated oncogenic transformation. Mol. Biol. Cell 2011, 22, 165–178. https://doi.org/10.1091/mbc.E10-06-0500.
- 62.Lee, Y.R.; Wu, S.Y.; Chen, R.Y.; Lin, Y.S.; Yeh, T.M.; Liu, H.S. Regulation of autophagy, glucose uptake, and glycolysis under dengue virus infection. Kaohsiung J. Med. Sci. 2020, 36, 911–919. https://doi.org/10.1002/kjm2.12271.
- 63.Dodson, M.; Darley-Usmar, V.; Zhang, J. Cellular metabolic and autophagic pathways: Traffic control by redox signaling. Free Radic. Biol. Med. 2013, 63, 207–221. https://doi.org/10.1016/j.freeradbiomed.2013.05.014.
- 64.Colell, A.; Ricci, J.E.; Tait, S.; Milasta, S.; Maurer, U.; Bouchier-Hayes, L.; Fitzgerald, P.; Guio-Carrion, A.; Waterhouse, N.J.; Li, C.W.; et al. GAPDH and autophagy preserve survival after apoptotic cytochrome c release in the absence of caspase activation. Cell 2007, 129, 983–997. https://doi.org/10.1016/j.cell.2007.03.045.
- 65.Craven, R.J.; Frazier, H.N.; Thibault, O. Dependence of glucose transport on autophagy and GAPDH activity. Brain Res. 2022, 1776, 147747. https://doi.org/10.1016/j.brainres.2021.147747.
- 66.Lee, M.N.; Ha, S.H.; Kim, J.; Koh, A.; Lee, C.S.; Kim, J.H.; Jeon, H.; Kim, D.H.; Suh, P.G.; Ryu, S.H. Glycolytic flux signals to mTOR through glyceraldehyde-3-phosphate dehydrogenase-mediated regulation of Rheb. Mol. Cell Biol. 2009, 29, 3991–4001. https://doi.org/10.1128/mcb.00165-09.
- 67.Yang, X.P.; Zheng, Y.Z.; Tan, J.; Tian, R.J.; Shen, P.; Cai, W.J.; Liao, H.Y. Circ_0020123 regulates autophagy, glycolysis, and malignancy by upregulating IRF4 through eliminating miR-193a-3p-mediated suppression of IRF4 in non-small cell lung cancer. Neoplasma 2022, 69, 392–403. https://doi.org/10.4149/neo_2022_211013N1449.
- 68.Fan, Q.; Yang, L.; Zhang, X.; Ma, Y.; Li, Y.; Dong, L.; Zong, Z.; Hua, X.; Su, D.; Li, H.; et al. Autophagy promotes metastasis and glycolysis by upregulating MCT1 expression and Wnt/β-catenin signaling pathway activation in hepatocellular carcinoma cells. J. Exp. Clin. Cancer Res. CR 2018, 37, 9. https://doi.org/10.1186/s13046-018-0673-y.
- 69.Li, T.; Tong, H.; Yin, H.; Luo, Y.; Zhu, J.; Qin, Z.; Yin, S.; He, W. Starvation induced autophagy promotes the progression of bladder cancer by LDHA mediated metabolic reprogramming. Cancer Cell Int. 2021, 21, 597. https://doi.org/10.1186/s12935-021-02303-1.
- 70.Wang, X.; Li, Z.; Gao, Z.; Li, Q.; Jiang, L.; Geng, C.; Yao, X.; Shi, X.; Liu, Y.; Cao, J. Cadmium induces cell growth in A549 and HELF cells via autophagy-dependent glycolysis. Toxicol. Vitr. 2020, 66, 104834. https://doi.org/10.1016/j.tiv.2020.104834.
- 71.Jeon, J.Y.; Lee, H.; Park, J.; Lee, M.; Park, S.W.; Kim, J.S.; Lee, M.; Cho, B.; Kim, K.; Choi, A.M.; et al. The regulation of glucose-6-phosphatase and phosphoenolpyruvate carboxykinase by autophagy in low-glycolytic hepatocellular carcinoma cells. Biochem. Biophys. Res. Commun. 2015, 463, 440–446. https://doi.org/10.1016/j.bbrc.2015.05.103.
- 72.Wang, H.J.; Park, J.Y.; Kwon, O.; Choe, E.Y.; Kim, C.H.; Hur, K.Y.; Lee, M.S.; Yun, M.; Cha, B.S.; Kim, Y.B.; et al. Chronic HMGCR/HMG-CoA reductase inhibitor treatment contributes to dysglycemia by upregulating hepatic gluconeogenesis through autophagy induction. Autophagy 2015, 11, 2089–2101. https://doi.org/10.1080/15548627.2015.1091139.
- 73.Fang, F.; Shi, X.; Brown, M.S.; Goldstein, J.L.; Liang, G. Growth hormone acts on liver to stimulate autophagy, support glucose production, and preserve blood glucose in chronically starved mice. Proc. Natl. Acad. Sci. USA 2019, 116, 7449–7454. https://doi.org/10.1073/pnas.1901867116.
- 74.Ezaki, J.; Matsumoto, N.; Takeda-Ezaki, M.; Komatsu, M.; Takahashi, K.; Hiraoka, Y.; Taka, H.; Fujimura, T.; Takehana, K.; Yoshida, M.; et al. Liver autophagy contributes to the maintenance of blood glucose and amino acid levels. Autophagy 2011, 7, 727–736. https://doi.org/10.4161/auto.7.7.15371.
- 75.Wang, X.P.; Huang, Z.; Li, Y.L.; Jin, K.Y.; Dong, D.J.; Wang, J.X.; Zhao, X.F. Krüppel-like factor 15 integrated autophagy and gluconeogenesis to maintain glucose homeostasis under 20-hydroxyecdysone regulation. PLoS Genet. 2022, 18, e1010229. https://doi.org/10.1371/journal.pgen.1010229.
- 76.Yang, Y.; Zhao, C.; Yang, P.; Wang, X.; Wang, L.; Chen, A. Autophagy in cardiac metabolic control: Novel mechanisms for cardiovascular disorders. Cell Biol. Int. 2016, 40, 944–954. https://doi.org/10.1002/cbin.10626.
- 77.Stephenson, M.C.; Leverton, E.; Khoo, E.Y.; Poucher, S.M.; Johansson, L.; Lockton, J.A.; Eriksson, J.W.; Mansell, P.; Morris, P.G.; MacDonald, I.A. Variability in fasting lipid and glycogen contents in hepatic and skeletal muscle tissue in subjects with and without type 2 diabetes: A 1H and 13C MRS study. NMR Biomed. 2013, 26, 1518–1526. https://doi.org/10.1002/nbm.2985.
- 78.Du, Q.; Wu, X.; Ma, K.; Liu, W.; Liu, P.; Hayashi, T.; Mizuno, K.; Hattori, S.; Fujisaki, H.; Ikejima, T. Silibinin alleviates ferroptosis of rat islet β cell INS-1 induced by the treatment with palmitic acid and high glucose through enhancing PINK1/parkin-mediated mitophagy. Arch. Biochem. Biophys. 2023, 743, 109644. https://doi.org/10.1016/j.abb.2023.109644.
- 79.Lei, J.; Alexander, A.S.; Pamela, S.; Chendong, Y.; Seth, J.P.; Qiong, A.W.; Lance, S.T.; Nicholas, D.A.; Michael, T.M.; Beth, P.; et al. Reductive carboxylation supports redox homeostasis during anchorage-independent growth. Nature 2016, 532, 255–258. https://doi.org/10.1038/nature17393.
- 80.Xiong, Z.; Yang, L.; Zhang, C.; Huang, W.; Zhong, W.; Yi, J.; Feng, J.; Zouxu, X.; Song, L.; Wang, X. MANF facilitates breast cancer cell survival under glucose-starvation conditions via PRKN-mediated mitophagy regulation. Autophagy 2025, 21, 80–101. https://doi.org/10.1080/15548627.2024.2392415.
- 81.Zhang, S.; Chen, S.; Sun, D.; Li, S.; Sun, J.; Gu, Q.; Liu, P.; Wang, X.; Zhu, H.; Xu, X.; et al. TIN2-mediated reduction of mitophagy induces RPE senescence under high glucose. Cell Signal 2024, 119, 111188. https://doi.org/10.1016/j.cellsig.2024.111188.
- 82.Wu, D.; Huang, W.; Zhang, J.; He, L.; Chen, S.; Zhu, S.; Sang, Y.; Liu, K.; Hou, G.; Chen, B.; et al. Downregulation of VEGFA accelerates AGEs-mediated nucleus pulposus degeneration through inhibiting protective mitophagy in high glucose environments. Int. J. Biol. Macromol. 2024, 262, 129950. https://doi.org/10.1016/j.ijbiomac.2024.129950.
- 83.Yang, W.; Qiu, C.; Lv, H.; Zhang, Z.; Yao, T.; Huang, L.; Wu, G.; Zhang, X.; Chen, J.; He, Y. Sirt3 Protects Retinal Pigment Epithelial Cells From High Glucose-Induced Injury by Promoting Mitophagy Through the AMPK/mTOR/ULK1 Pathway. Transl. Vis. Sci. Technol. 2024, 13, 19. https://doi.org/10.1167/tvst.13.3.19.
- 84.Lee, H.; Cho, S.; Kim, M.J.; Park, Y.J.; Cho, E.; Jo, Y.S.; Kim, Y.S.; Lee, J.Y.; Thoudam, T.; Woo, S.H.; et al. ApoE4-dependent lysosomal cholesterol accumulation impairs mitochondrial homeostasis and oxidative phosphorylation in human astrocytes. Cell Rep. 2023, 42, 113183. https://doi.org/10.1016/j.celrep.2023.113183.
- 85.Wang, W.; Zhang, Y.; Huang, W.; Yuan, Y.; Hong, Q.; Xie, Z.; Li, L.; Chen, Y.; Li, X.; Meng, Y. Alamandine/MrgD axis prevents TGF-β1-mediated fibroblast activation via regulation of aerobic glycolysis and mitophagy. J. Transl. Med. 2023, 21, 24. https://doi.org/10.1186/s12967-022-03837-2.
- 86.Dai, T.; Zhang, X.; Li, M.; Tao, X.; Jin, M.; Sun, P.; Zhou, Q.; Jiao, L. Dietary vitamin K(3) activates mitophagy, improves antioxidant capacity, immunity and affects glucose metabolism in Litopenaeus vannamei. Food Funct. 2022, 13, 6362–6372. https://doi.org/10.1039/d2fo00865c.
- 87.Li, S.; Wang, Y.; Zhang, X.; Xiong, X.; Zhou, F.; Li, X.; Fan, J.; Liang, X.; Li, G.; Peng, Y.; et al. Mitochondrial damage-induced abnormal glucose metabolism with ageing in the hippocampus of APP/PS1 mice. Metabolomics 2023, 19, 56. https://doi.org/10.1007/s11306-023-02023-9.
- 88.Li, Z.; Meng, X.; Ma, G.; Liu, W.; Li, W.; Cai, Q.; Wang, S.; Huang, G.; Zhang, Y. Increasing brain glucose metabolism by ligustrazine piperazine ameliorates cognitive deficits through PPARγ-dependent enhancement of mitophagy in APP/PS1 mice. Alzheimers Res. Ther. 2022, 14, 150. https://doi.org/10.1186/s13195-022-01092-7.
- 89.Su, D.; Song, Y.; Li, D.; Yang, S.; Zhan, S.; Zhong, T.; Guo, J.; Cao, J.; Li, L.; Zhang, H.; et al. Cold exposure affects glucose metabolism, lipid droplet deposition and mitophagy in skeletal muscle of newborn goats. Domest. Anim. Endocrinol. 2024, 88, 106847. https://doi.org/10.1016/j.domaniend.2024.106847.
- 90.Li, Y.; Gao, Y.; Yu, G.; Ye, Y.; Zhu, H.; Wang, J.; Li, Y.; Chen, L.; Gu, L. G6PD protects against cerebral ischemia-reperfusion injury by inhibiting excessive mitophagy. Life Sci. 2025, 362, 123367. https://doi.org/10.1016/j.lfs.2024.123367.
- 91.Yuan, M.; Yao, Y.; Wu, D.; Zhu, C.; Dong, S.; Tong, X. Pannexin1 inhibits autophagy of cisplatin-resistant testicular cancer cells by mediating ATP release. Cell Cycle 2022, 21, 1651–1661. https://doi.org/10.1080/15384101.2022.2060655.
- 92.Junfang, Y.; Yi, X.; Fang, W.; Yuhong, C.; Jinhua, Z.; Zhihui, D.; Lu, G.; Hongyan, L.; Jing, S.; Chao, S.; et al. Carbon ion combined with tigecycline inhibits lung cancer cell proliferation by inducing mitochondrial dysfunction. Life Sci. 2020, 263, 118586. https://doi.org/10.1016/j.lfs.2020.118586.
- 93.Yidan, W.; Isabelle, M.; Yuting, M.; Oliver, K.; Lorenzo, G.; Guido, K. Autophagy-dependent ATP release from dying cells via lysosomal exocytosis. Autophagy 2013, 9, 1624–1625. https://doi.org/10.4161/auto.25873.
- 94.Michaud, M.; Martins, I.; Sukkurwala, A.; Adjemian, S.; Ma, Y.; Pellegatti, P.; Shen, S.; Kepp, O.; Scoazec, M.; Mignot, G.; et al. Autophagy-dependent anticancer immune responses induced by chemotherapeutic agents in mice. Science 2011, 334, 1573–1577. https://doi.org/10.1126/science.1208347.
- 95.Peynshaert, K.; Manshian, B.; Joris, F.; Braeckmans, K.; De Smedt, S.; Demeester, J.; Soenen, S. Exploiting intrinsic nanoparticle toxicity: The pros and cons of nanoparticle-induced autophagy in biomedical research. Chem. Rev. 2014, 114, 7581–7609. https://doi.org/10.1021/cr400372p.
- 96.Naoki, I.; Urs, T.R.; Shin’ichi, T. ATP-Induced Increase in Intracellular Calcium Levels and Subsequent Activation of mTOR as Regulators of Skeletal Muscle Hypertrophy. Int. J. Mol. Sci. 2018, 19, 2804. https://doi.org/10.3390/ijms19092804.
- 97.Bian, S.; Sun, X.; Bai, A.; Zhang, C.; Li, L.; Enjyoji, K.; Junger, W.G.; Robson, S.C.; Wu, Y. P2X7 integrates PI3K/AKT and AMPK-PRAS40-mTOR signaling pathways to mediate tumor cell death. PLoS ONE 2013, 8, e60184–e60184. https://doi.org/10.1371/journal.pone.0060184.
- 98.Jiang, Y.; Lin, J.; Zheng, H.; Zhu, P. The Role of Purinergic Signaling in Heart Transplantation. Front. Immunol. 2022, 13, 826943. https://doi.org/10.3389/fimmu.2022.826943.
- 99.Yin, H.; Tang, X.; Peng, Y.; Wen, H.; Yang, H.; Li, S.; Zheng, X.; Xiong, Y. Pannexin-1 regulation of ATP release promotes the invasion of pituitary adenoma. J. Endocrinol. Investig. 2025, 48, 317–332. https://doi.org/10.1007/s40618-024-02445-9.
- 100.Bartlett, P.J.; Gaspers, L.D.; Pierobon, N.; Thomas, A.P. Calcium-dependent regulation of glucose homeostasis in the liver. Cell Calcium 2014, 55, 306–316. https://doi.org/10.1016/j.ceca.2014.02.007.
- 101.Kania, E.; Pająk, B.; Orzechowski, A. Calcium homeostasis and ER stress in control of autophagy in cancer cells. Biomed. Res. Int. 2015, 2015, 352794. https://doi.org/10.1155/2015/352794.
- 102.Høyer-Hansen, M.; Jäättelä, M. Connecting endoplasmic reticulum stress to autophagy by unfolded protein response and calcium. Cell Death Differ. 2007, 14, 1576–1582. https://doi.org/10.1038/sj.cdd.4402200.
- 103.Kondratskyi, A.; Yassine, M.; Kondratska, K.; Skryma, R.; Slomianny, C.; Prevarskaya, N. Calcium-permeable ion channels in control of autophagy and cancer. Front. Physiol. 2013, 4, 272. https://doi.org/10.3389/fphys.2013.00272.
- 104.Cárdenas, C.; Miller, R.A.; Smith, I.; Bui, T.; Molgó, J.; Müller, M.; Vais, H.; Cheung, K.H.; Yang, J.; Parker, I.; et al. Essential regulation of cell bioenergetics by constitutive InsP3 receptor Ca2+ transfer to mitochondria. Cell 2010, 142, 270–283. https://doi.org/10.1016/j.cell.2010.06.007.
- 105.Yuan, Y.H.; Yan, W.F.; Sun, J.D.; Huang, J.Y.; Mu, Z.; Chen, N.H. The molecular mechanism of rotenone-induced α-synuclein aggregation: Emphasizing the role of the calcium/GSK3β pathway. Toxicol. Lett. 2015, 233, 163–171. https://doi.org/10.1016/j.toxlet.2014.11.029.
- 106.Hoyer-Hansen, M.; Bastholm, L.; Szyniarowski, P.; Campanella, M.; Szabadkai, G.; Farkas, T.; Bianchi, K.; Fehrenbacher, N.; Elling, F.; Rizzuto, R.; et al. Control of macroautophagy by calcium, calmodulin-dependent kinase kinase-beta, and Bcl-2. Mol. Cell 2007, 25, 193–205. https://doi.org/10.1016/j.molcel.2006.12.009.
- 107.Shi, W.; Xu, D.; Gu, J.; Xue, C.; Yang, B.; Fu, L.; Song, S.; Liu, D.; Zhou, W.; Lv, J.; et al. Saikosaponin-d inhibits proliferation by up-regulating autophagy via the CaMKKβ-AMPK-mTOR pathway in ADPKD cells. Mol. Cell. Biochem. 2018, 449, 219–226. https://doi.org/10.1007/s11010-018-3358-0.
- 108.Félix-Martínez, G.J.; Godínez-Fernández, J.R. Mathematical models of electrical activity of the pancreatic β-cell: A physiological review. Islets 2014, 6, e949195. https://doi.org/10.4161/19382014.2014.949195.
- 109.Zhang, N.; Liu, F.; Zhao, Y.; Sun, X.; Wen, B.; Lu, J.Q.; Yan, C.; Li, D. Defect in degradation of glycogenin-exposed residual glycogen in lysosomes is the fundamental pathomechanism of Pompe disease. J. Pathol. 2024, Online ahead of print. https://doi.org/10.1002/path.6255.
- 110.Kalamidas, S.A.; Kotoulas, O.B.; Hann, A.C. Studies on glycogen autophagy: Effects of phorbol myristate acetate, ionophore A23187, or phentolamine. Microsc. Res. Tech. 2002, 57, 507–511. https://doi.org/10.1002/jemt.10104.
- 111.Santoni, G.; Santoni, M.; Nabissi, M. Functional role of T-type calcium channels in tumour growth and progression: Prospective in cancer therapy. Br. J. Pharmacol. 2012, 166, 1244–1246. https://doi.org/10.1111/j.1476-5381.2012.01908.x.
- 112.Andrea, W.; Sovan, S.; Paul, C.; Evangelia, K.T.; Shinji, S.; Farah, H.S.; Luca, J.; Angeleen, F.; Dean, P.; Paul, G.; et al. Novel targets for Huntington’s disease in an mTOR-independent autophagy pathway. Nat. Chem. Biol. 2008, 4, 295–305. https://doi.org/10.1038/nchembio.79.
- 113.Busch Sørensen, M.; Sjøstrand, H.; Sengeløv, H.; Tiefenthal Thrane, M.; Juul Holst, J.; Lyngsøe, J. Influence of short term verapamil treatment on glucose metabolism in patients with non-insulin dependent diabetes mellitus. Eur. J. Clin. Pharmacol. 1991, 41, 401–404. https://doi.org/10.1007/bf00626359.
- 114.Ardizzone, T.D.; Lu, X.H.; Dwyer, D.S. Calcium-independent inhibition of glucose transport in PC-12 and L6 cells by calcium channel antagonists. Am. J. Physiol. Cell Physiol. 2002, 283, C579–C586. https://doi.org/10.1152/ajpcell.00451.2001.
- 115.Pajak, B.; Kania, E.; Gajkowska, B.; Orzechowski, A. Verapamil-induced autophagy-like process in colon adenocarcinoma COLO 205 cells; the ultrastructural studies. Pharmacol. Rep. 2012, 64, 991–996. https://doi.org/10.1016/s1734-1140(12)70896-4.
- 116.Tomar, D.; Elrod, J.W. Metabolite regulation of the mitochondrial calcium uniporter channel. Cell Calcium 2020, 92, 102288. https://doi.org/10.1016/j.ceca.2020.102288.
- 117.Gray, L.R.; Sultana, M.R.; Rauckhorst, A.J.; Oonthonpan, L.; Tompkins, S.C.; Sharma, A.; Fu, X.; Miao, R.; Pewa, A.D.; Brown, K.S.; et al. Hepatic Mitochondrial Pyruvate Carrier 1 Is Required for Efficient Regulation of Gluconeogenesis and Whole-Body Glucose Homeostasis. Cell Metab. 2015, 22, 669–681. https://doi.org/10.1016/j.cmet.2015.07.027.
- 118.Nemani, N.; Dong, Z.; Daw, C.C.; Madaris, T.R.; Ramachandran, K.; Enslow, B.T.; Rubannelsonkumar, C.S.; Shanmughapriya, S.; Mallireddigari, V.; Maity, S.; et al. Mitochondrial pyruvate and fatty acid flux modulate MICU1-dependent control of MCU activity. Sci. Signal 2020, 13, eaaz6206. https://doi.org/10.1126/scisignal.aaz6206.
- 119.Fan, Y.; Wang, N.; Rocchi, A.; Zhang, W.; Vassar, R.; Zhou, Y.; He, C. Identification of natural products with neuronal and metabolic benefits through autophagy induction. Autophagy 2017, 13, 41–56. https://doi.org/10.1080/15548627.2016.1240855.
- 120.Goginashvili, A.; Zhang, Z.; Erbs, E.; Spiegelhalter, C.; Kessler, P.; Mihlan, M.; Pasquier, A.; Krupina, K.; Schieber, N.; Cinque, L.; et al. Insulin granules. Insulin secretory granules control autophagy in pancreatic β cells. Science 2015, 347, 878–882. https://doi.org/10.1126/science.aaa2628.
- 121.Rutter, G.A. Cell biology. Pancreas micromanages autophagy. Science 2015, 347, 826–827. https://doi.org/10.1126/science.aaa
- 122.Yamamoto, S.; Kuramoto, K.; Wang, N.; Situ, X.; Priyadarshini, M.; Zhang, W.; Cordoba-Chacon, J.; Layden, B.; He, C. Autophagy Differentially Regulates Insulin Production and Insulin Sensitivity. Cell Rep. 2018, 23, 3286–3299. https://doi.org/10.1016/j.celrep.2018.05.032.
- 123.Bartolome, A.; Guillen, C.; Benito, M. Autophagy plays a protective role in endoplasmic reticulum stress-mediated pancreatic β cell death. Autophagy 2012, 8, 1757–1768. https://doi.org/10.4161/auto.21994.
- 124.Sheng, Q.; Xiao, X.; Prasadan, K.; Chen, C.; Ming, Y.; Fusco, J.; Gangopadhyay, N.N.; Ricks, D.; Gittes, G.K. Autophagy protects pancreatic beta cell mass and function in the setting of a high-fat and high-glucose diet. Sci. Rep. 2017, 7, 16348. https://doi.org/10.1038/s41598-017-16485-0.
- 125.Bachar-Wikstrom, E.; Wikstrom, J.D.; Ariav, Y.; Tirosh, B.; Kaiser, N.; Cerasi, E.; Leibowitz, G. Stimulation of autophagy improves endoplasmic reticulum stress-induced diabetes. Diabetes 2013, 62, 1227–1237. https://doi.org/10.2337/db12-1474.
- 126.Wang, P.; Li, J.; Li, C.G.; Zhou, X.; Chen, X.; Zhu, M.; Wang, H. Restoring Autophagy by Exercise Ameliorates Insulin Resistance Partly via Calcineurin-Driven TFEB Nuclear Translocation. Clin. Exp. Pharmacol. Physiol. 2025, 52, e70010. https://doi.org/10.1111/1440-1681.70010.
- 127.Luan, B.; Sun, C. MiR-138-5p affects insulin resistance to regulate type 2 diabetes progression through inducing autophagy in HepG2 cells by regulating SIRT1. Nutr. Res. 2018, 59, 90–98. https://doi.org/10.1016/j.nutres.2018.05.001.
- 128.Kang, Y.H.; Cho, M.H.; Kim, J.Y.; Kwon, M.S.; Peak, J.J.; Kang, S.W.; Yoon, S.Y.; Song, Y. Impaired macrophage autophagy induces systemic insulin resistance in obesity. Oncotarget 2016, 7, 35577–35591. https://doi.org/10.18632/oncotarget.9590.
- 129.Cai, J.; Pires, K.M.; Ferhat, M.; Chaurasia, B.; Buffolo, M.A.; Smalling, R.; Sargsyan, A.; Atkinson, D.L.; Summers, S.A.; Graham, T.E.; et al. Autophagy Ablation in Adipocytes Induces Insulin Resistance and Reveals Roles for Lipid Peroxide and Nrf2 Signaling in Adipose-Liver Crosstalk. Cell Rep. 2018, 25, 1708–1717.e1705. https://doi.org/10.1016/j.celrep.2018.10.040.
- 130.Rovira-Llopis, S.; Diaz-Morales, N.; Banuls, C.; Blas-Garcia, A.; Polo, M.; Lopez-Domenech, S.; Jover, A.; Rocha, M.; Hernandez-Mijares, A.; Victor, V.M. Is Autophagy Altered in the Leukocytes of Type 2 Diabetic Patients? Antioxid. Redox Signal. 2015, 23, 1050–1056. https://doi.org/10.1089/ars.2015.6447.
- 131.Song, R.; Zhao, X.; Cao, R.; Liang, Y.; Zhang, D.Q.; Wang, R. Irisin improves insulin resistance by inhibiting autophagy through the PI3K/Akt pathway in H9c2 cells. Gene 2021, 769, 145209. https://doi.org/10.1016/j.gene.2020.145209.
- 132.Kim, K.H.; Jeong, Y.T.; Oh, H.; Kim, S.H.; Cho, J.M.; Kim, Y.N.; Kim, S.S.; Kim, D.H.; Hur, K.Y.; Kim, H.K.; et al. Autophagy deficiency leads to protection from obesity and insulin resistance by inducing Fgf21 as a mitokine. Nat. Med. 2013, 19, 83–92. https://doi.org/10.1038/nm.3014.
- 133.Li, S.; Li, H.; Yang, D.; Yu, X.; Irwin, D.M.; Niu, G.; Tan, H. Excessive Autophagy Activation and Increased Apoptosis Are Associated with Palmitic Acid-Induced Cardiomyocyte Insulin Resistance. J. Diabetes Res. 2017, 2017, 2376893. https://doi.org/10.1155/2017/2376893.
- 134.Feng, J.; Li, J.; Wu, L.; Yu, Q.; Ji, J.; Wu, J.; Dai, W.; Guo, C. Emerging roles and the regulation of aerobic glycolysis in hepatocellular carcinoma. J. Exp. Clin. Cancer Res. CR 2020, 39, 126. https://doi.org/10.1186/s13046-020-01629-4.
- 135.Prieto-Dominguez, N.; Ordonez, R.; Fernandez, A.; Garcia-Palomo, A.; Muntane, J.; Gonzalez-Gallego, J.; Mauriz, J.L. Modulation of Autophagy by Sorafenib: Effects on Treatment Response. Front. Pharmacol. 2016, 7, 151. https://doi.org/10.3389/fphar.2016.00151.
- 136.Yan, X.; Tian, R.; Sun, J.; Zhao, Y.; Liu, B.; Su, J.; Li, M.; Sun, W.; Xu, X. Sorafenib-Induced Autophagy Promotes Glycolysis by Upregulating the p62/HDAC6/HSP90 Axis in Hepatocellular Carcinoma Cells. Front. Pharmacol. 2021, 12, 788667. https://doi.org/10.3389/fphar.2021.788667.
- 137.Cao, B.; Deng, H.; Cui, H.; Zhao, R.; Li, H.; Wei, B.; Chen, L. Knockdown of PGM1 enhances anticancer effects of orlistat in gastric cancer under glucose deprivation. Cancer Cell Int. 2021, 21, 481. https://doi.org/10.1186/s12935-021-02193-3.
- 138.Peng, H.; Wang, Q.; Qi, X.; Wang, X.; Zhao, X. Orlistat induces apoptosis and protective autophagy in ovarian cancer cells: Involvement of Akt-mTOR-mediated signaling pathway. Arch. Gynecol. Obstet. 2018, 298, 597–605. https://doi.org/10.1007/s00404-018-4841-2.
- 139.Sun, B.; Ou, H.; Ren, F.; Huan, Y.; Zhong, T.; Gao, M.; Cai, H.J.M.m. Propofol inhibited autophagy through Ca/CaMKKβ/AMPK/mTOR pathway in OGD/R-induced neuron injury. Mol. Med. 2018, 24, 58. https://doi.org/10.1186/s10020-018-0054-1.
- 140.Li, A.; Yi, B.; Han, H.; Yang, S.; Hu, Z.; Zheng, L.; Wang, J.; Liao, Q.; Zhang, H. Vitamin D-VDR (vitamin D receptor) regulates defective autophagy in renal tubular epithelial cell in streptozotocin-induced diabetic mice via the AMPK pathway. Autophagy 2022, 18, 877–890. https://doi.org/10.1080/15548627.2021.1962681.
- 141.Rim, H.K.; Cho, S.; Shin, D.H.; Chung, K.S.; Cho, Y.W.; Choi, J.H.; Lee, J.Y.; Lee, K.T. T-type Ca2+ channel blocker, KYS05090 induces autophagy and apoptosis in A549 cells through inhibiting glucose uptake. Molecules 2014, 19, 9864–9875. https://doi.org/10.3390/molecules19079864.
How to Cite
Liu, M.; Luo, X.; Hu, H.; Chen, L. Autophagy: A Major Regulatory Factor in Glucose Metabolism. Health and Metabolism 2025, 2 (4), 1. https://doi.org/10.53941/hm.2025.100024.
RIS
BibTex
Copyright & License

Copyright (c) 2025 by the authors.
This work is licensed under a Creative Commons Attribution 4.0 International License.
Contents
References


